- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assessment of SREBP Activation Using a Microsomal Vesicle Budding Assay
Published: Vol 14, Iss 24, Dec 20, 2024 DOI: 10.21769/BioProtoc.5139 Views: 1567
Reviewed by: Philipp A.M. SchmidpeterShailesh KumarMaria Falzone

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Novel Imaging Technique for The On-site Assessment of Renal Biopsy Specimens
Tomoaki Takata [...] Kentaro Yamada
Sep 20, 2022 1743 Views

In-Gel Activity Assay of Mammalian Mitochondrial and Cytosolic Aconitases, Surrogate Markers of Compartment-Specific Oxidative Stress and Iron Status
Wing-Hang Tong and Tracey A. Rouault
Dec 5, 2024 2220 Views

Proteome Birthdating: A Single-Sample Approach for Measuring Global Turnover Dynamics and “Protein Age”
Michael E. Meadow [...] Sina Ghaemmaghami
May 5, 2025 2222 Views
Abstract
Sterol regulatory element binding proteins (SREBPs) are transcription factors that reside in the endoplasmic reticulum (ER) membrane as inactive precursors. To be active, SREBPs are translocated to the Golgi where the transcriptionally active N-terminus is cleaved and released to the nucleus to regulate gene expression. Nuclear SREBP levels can be determined by immunoblot analysis; however, this method can only determine the steady-state levels of nuclear SREBPs and does not capture the actual status of activation. The vesicle budding assay provides an alternative way to quantify the activation of SREBPs by monitoring the initiation of SREBP translocation from the ER to the Golgi through vesicles. Microsomal membranes isolated from the liver are incubated in a reaction buffer containing the necessary components to facilitate vesicle formation. Microsomal membranes and vesicles are isolated and SREBPs are quantified in each by immunoblot analysis. The amount of SREBPs found in the budded vesicles provides an assessment of the SREBP activation in the liver.
Key features
• This protocol describes a method to isolate budding vesicles from liver ER membranes
• The in vitro budding assay can be applied to investigate the movement of proteins from the ER to the Golgi
• This protocol was developed based on the procedures described previously with cultured cells [1–3]
Keywords: Endoplasmic reticulum (ER)Graphical overview
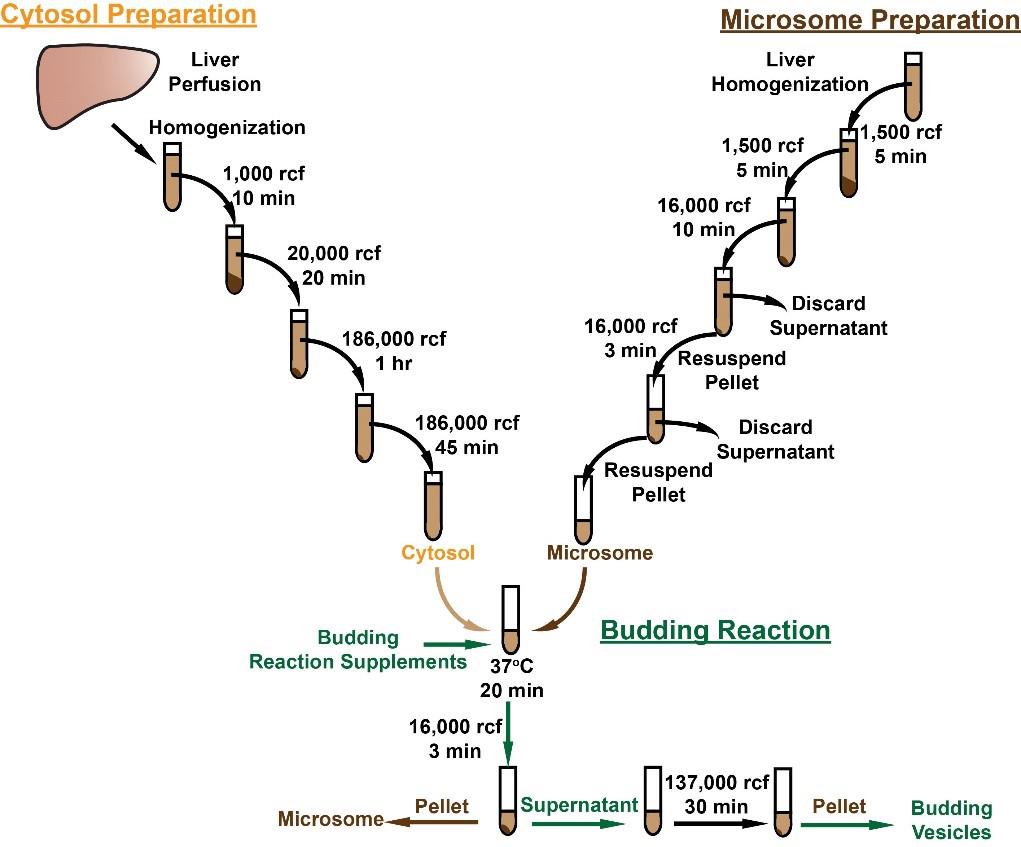
Background
The transport of proteins from the endoplasmic reticulum (ER) to the Golgi complex is required for additional protein modifications and/or protein secretion. Small transport vesicles that bud from mammalian ER and fuse to Golgi were reported in the late 1980s [4] and further characterized thereafter [2,3,5]. Sterol regulatory element binding proteins (SREBPs) are a family of membrane-bound transcription factors that regulate the expression of genes involved in cholesterol and fatty acid homeostasis [6–8], which utilize small vesicles for transport from the ER to the Golgi for activation [1]. Newly synthesized SREBP proteins are embedded in the ER membrane, forming a complex with SREBP cleavage activating protein (SCAP), a protein that serves as a sterol sensor and escort protein. The process of SREBP activation requires the membrane-bound SREBP/SCAP complex to move from the ER to Golgi, where two proteases reside, which cleave and free the transcriptionally active N-terminal portion of SREBPs from the membrane. The N-terminal SREBPs then enter the nucleus and regulate the transcription of downstream genes [8].
Immunoblot analysis is routinely used to evaluate the degree of SREBP activation. The molecular weight of the cleaved mature/nuclear form of SREBPs is ~68 kD, while the molecular weight of the membrane-bound precursor SREBP is ~125 kD. By quantifying the amount of cleaved nuclear SREBPs, an estimation of SREBP activation can be achieved. However, nuclear SREBP levels represent a steady-state measurement of the combined results of both protein activation and degradation. In a recent study, we developed a new method to determine SREBP activation by measuring the full-length SREBPs in budding vesicles of mouse liver. Although the in vitro budding reaction might have the limitation of not reflecting the in vivo status, this method measures the initiation of SREBP transportation from the ER to Golgi for cleavage activation. Compared to measuring the cleaved nuclear form SREBPs, evaluation of SREBP activation by measuring the SREBPs in the budding vesicles eliminates the potential effects of degradation of nuclear SREBPs.
Materials and reagents
Biological materials
Note: The general microsomal budding assay is not dependent on the animal or diet used.
Human SREBP-1c transgenic rat
High carbohydrate diet (MP Biomedicals, catalog number: 960238)
HEPES (Sigma-Aldrich, catalog number: H3375)
D-Sorbitol (Sigma-Aldrich, catalog number: S6021)
Potassium acetate (Sigma-Aldrich, catalog number: P1190)
Magnesium acetate tetrahydrate (Sigma-Aldrich, catalog number: M5661)
0.5 M EGTA-KOH, pH 8.0 (Fisher Scientific, catalog number: 50-997-744)
1 M Tris-Cl, pH 6.8 (Fisher Scientific, catalog number: 50-843-263)
Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D9779)
Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: 75746)
Bromophenol blue (Sigma-Aldrich, catalog number: B0126)
Glycerol (Sigma-Aldrich, catalog number: G5516)
Trizma base (Sigma-Aldrich, catalog number: T1503)
Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P8340)
0.9% Sodium chloride irrigation (Saline) (Baxter, catalog number: BX-2F7124)
Isoflurane (Piramal Critical Care, catalog number: 33794-013-25)
Creatine phosphate (Millipore Sigma, catalog number: 10621714001)
ATP (Sigma-Aldrich, catalog number: G6419)
GTP (Sigma-Aldrich, catalog number: G8877)
Creatine kinase (Millipore Sigma, catalog number: 10127566001)
Anti-HA antibody (Cell Signaling, catalog number: 3724)
Anti-Calnexin antibody (Enzo, catalog number: ADI-SPA-860-F)
Anti-ERGIC-53 antibody (Abcam, catalog number: ab125006)
Goat anti-rabbit IgG, F(ab’)2 (Jackson Immuno, catalog number: 115-035-072)
PierceTM BCA Protein Assay kit (Thermo Fisher, catalog number: A65453)
SuperSignal West Pico chemiluminescent substrate (Thermo Fisher, catalog number: 23225)
Buffer 1 (see Recipes)
Buffer 2 (see Recipes)
5× SDS-loading buffer (see Recipes)
20× budding reaction supplement (see Recipes)
Recipes
pH values of buffers are all adjusted at room temperature.
Buffer 1
Reagent Final concentration Quantity or Volume 1 M HEPES-KOH, pH 7.2 10 mM 500 μL 2 M Sorbitol 250 mM 6.25 mL 1 M KOAc 10 mM 500 µL 1 M Mg(OAc)2 1.5 mM 75 µL Total n/a Adjust pH to 7.2 with KOH, then add H2O to 50 mL Can be stored at 4 °C for up to two months.
Buffer 2
Reagent Final concentration Volume 1 M HEPES-KOH, pH 7.2 50 mM 2.5 mL 2 M Sorbitol 250 mM 6.25 mL 1 M KOAc 70 mM 3.5 mL 1 M Mg(OAc)2 2.5 mM 125 µL 0.5 M EGTA-KOH, pH 8.0 5 mM 500 µL Total n/a Adjust pH to 7.2 with KOH, then add H2O to 50 mL Can be stored at 4 °C for up to two months.
5× SDS-loading buffer
Reagent Final concentration Volume SDS 15% 15 g Bromophenol blue 0.02% 20 mg Glycerol 25% 25 mL 1 M Tris-Cl, pH 6.8 0.15 M 15 mL Total n/a Add H2O to 100 mL Add 62.5 μL of β-mercaptoethanol for each 500 μL of loading buffer freshly before use.
20× budding reaction supplement
The following reagents should be freshly prepared individually. Prepare the 20× budding reaction supplement by mixing equal volumes of each reagent immediately before carrying out the budding reaction.
Note: The molecular weight or specific activity of each reagent will be different depending on the source and lot number of the reagents. The quantity needs to be calculated based on the information from the vendor.
Reagent 20× concentration Quantity Final volume in H2O Creatine phosphate 200 mM 26.2 mg 100 µL ATP 30 mM 6.61 mg 100 µL GTP 10 mM 2.09 mg 100 µL Creatine kinase 80 U/mL 1 mg 10 mL
Laboratory supplies
18 G catheter (BD, catalog number: 382644)
Protein LoBind tubes (Eppendorf, catalog number: 022431081)
Polycarbonate centrifuge tubes (for AT3 rotor) (Beckman Coulter, catalog number: 343776)
Polycarbonate centrifuge tubes (for AT6 rotor) (Beckman Coulter, catalog number: 362305)
Equipment
Tabletop centrifuge (Eppendorf, model: Centrifuge 5424)
37 °C incubator (ThermoMixer, Eppendorf, model: F1.5 was used in this experiment)
Ultracentrifuge (Thermo Scientific, model: Sorvall MX120)
S100-AT6 fixed angle rotor (Thermo Scientific, catalog number: 45588)
S120-AT3 fixed angle rotor (Thermo Scientific, catalog number: 45584)
Peristaltic pump (Amersham Biosciences, model: Pump P-1)
Dounce tissue homogenizer-2 mL (DWK life sciences, catalog number: 885300-002)
Dounce tissue homogenizer-7 mL (DWK life sciences, catalog number: 885300-007)
Procedure
Preparation of cytosol from rat liver
Note: The rat used for cytosol preparation can be different from the animal used for microsomal preparation in section B. Cytosol can be prepared and aliquoted at -80 °C for long-term storage.
Fill the liver perfusion tube with ~100 mL of 0.9% saline and connect one end of the bubble trap tubular to the peristaltic pump and the other end to an 18 G catheter.
Euthanize the rat in an anesthetization jar filled with isoflurane.
Place the rat on a procedure pad, expose the abdominal cavity, and move the liver to expose the portal vein and the inferior vena cava (IVC).
Use an 18 G catheter to cannulate the portal vein; then, cut the IVC using scissors, start the perfusion pump at a rate of ~5 mL/min, and perfuse for 10 min (Figure 1A).
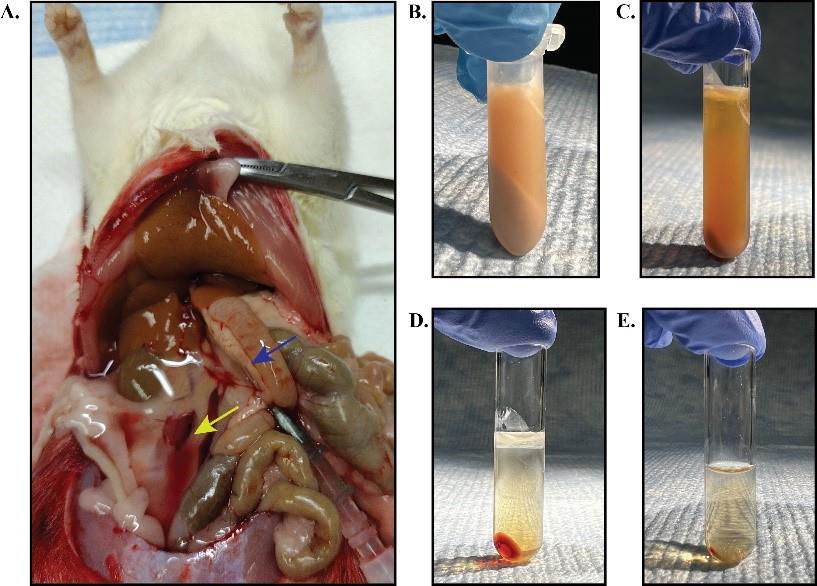
Figure 1. Preparation of cytosol from rat liver. A. Liver perfusion with 0.9% saline. The blue arrow indicates the portal vein location where the catheter was inserted. The yellow arrow shows the location where the IVC was cut. B–E. Samples after each centrifugation in the cytosol preparation steps. Lipid fractions (top) and pellets were discarded after each centrifugation. The animal experiments were performed with the approval of the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center.Remove the perfused liver and homogenize 2 g of liver in 5 mL of ice-cold buffer 2 supplemented with 1mM DTT using a 7 mL glass Dounce tissue homogenizer.
Centrifuge the homogenate at 4 °C as detailed below.
1,000 RCF for 10 min with a tabletop centrifuge. Remove the lipid fraction that floats on the supernatant with a pipette and only collect the clear fraction between the lipid layer and the pellet (Figure 1B). Transfer the clear supernatant to a polycarbonate centrifuge tube.
20,000 RCF for 20 min in an AT6 rotor using an ultracentrifuge machine; transfer the clear supernatant to a new polycarbonate centrifuge tube (Figure 1C).
186,000 RCF for 1 h in an AT6 rotor using an ultracentrifuge machine; transfer the clear supernatant to a new polycarbonate centrifuge tube (Figure 1D).
186,000 RCF for 45 min in an AT6 rotor using an ultracentrifuge machine (Figure 1E).
Collect the supernatant from the last spin (cytosol) and determine the protein concentration using a BCA Protein Assay kit. Aliquot the cytosol to individual tubes and store at -80 °C for future use.
Microsomal membrane isolation from liver
This protocol has been validated with mouse and rat livers.
Homogenize 200 mg of fresh liver in 1 mL of ice-cold buffer 1 supplemented with a protease inhibitor cocktail (100×) using a 2 mL Dounce homogenizer.
Centrifuge the liver homogenate at 1,500 RCF for 5 min in a tabletop centrifuge at 4 °C. Transfer the supernatant to a new tube and discard the pellet (Figure 2A).
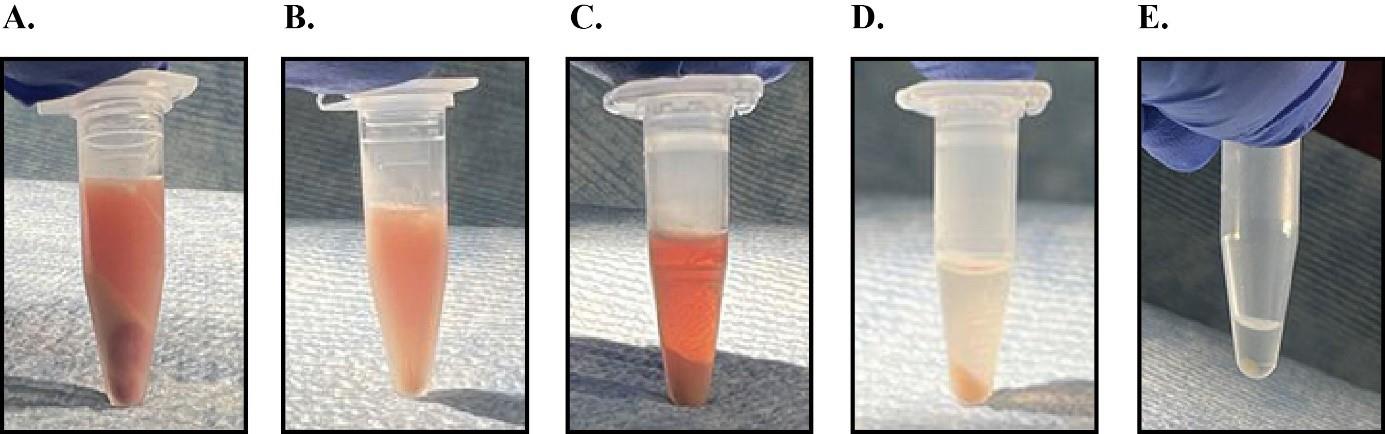
Figure 2. Samples after centrifugation during microsomal membrane preparations. Panels A–E show the outcomes after centrifugation in steps B2, B4, B5, B7, and C3 respectively.Centrifuge the supernatant from step B2 at 1,500 RCF for another 5 min at 4 °C.
Transfer the supernatant to a 1.5 mL protein low-bind tube. Avoid disturbing the remaining small pellet at the bottom (Figure 2B).
Centrifuge the supernatant at 16,000 RCF for 10 min in a tabletop centrifuge at 4 °C and remove and discard the supernatant (Figure 2C).
Resuspend the pellet in 400 µL of ice-cold buffer 2 supplemented with a protease inhibitor cocktail (100×) and transfer to a new protein low-bind tube.
Centrifuge the resuspended pellet at 16,000 RCF for 3 min in a tabletop centrifuge at 4 °C and remove and discard the supernatant (Figure 2D).
Resuspend the pellet (microsomal membranes) in 70 μL of ice-cold buffer 2 supplemented with the protease inhibitor cocktail (100×).
Determine the microsomal membrane protein concentration using the BCA Protein Assay kit.
In vitro budding reaction
For each reaction, add 80 μg of microsomal protein, 4 μL of the 20× budding reaction supplement, 600 μg of liver cytosol, and adjust the final reaction volume to 80 μL with buffer 2. Mix the reaction by pipetting up and down 10 times.
Incubate the reaction at 37 °C for 20 min, then transfer to ice for 5 min.
Centrifuge the reaction from step C2 at 16,000 RCF in a tabletop centrifuge for 3 min at 4 °C (Figure 2E).
Budding vesicle collection:
Transfer 65 μL of the supernatant from step C3 to a polycarbonate centrifuge tube and centrifuge at 137,000 RCF for 30 min at 4 °C in an AT3 rotor using an ultracentrifuge machine to isolate the formed vesicles.
Remove the supernatant, resuspend the pellet in 30 μL of buffer 2 supplemented with 5× SDS-loading buffer, and incubate at 55 °C for 20 min. Following the incubation, load the entire sample to an 8% SDS-PAGE for immunoblot analysis of proteins of interest that reside in the isolated budding vesicles.
Microsomal membrane collection: Resuspend the pellet (microsomal membrane) from step C3 in 60 μL of buffer 2 supplemented with 5× SDS-loading buffer and incubate at 55 °C for 20 min. Following the incubation, load 15 μL of the sample onto an 8% SDS-PAGE for immunoblot analysis.
Note: You might only see a very tiny pellet following the centrifugation in step C5. Mark the tube position in the rotor to help locate the pellet after centrifugation.
Validation of protocol
This protocol has been used and validated in the following research article:
Rong et al. [8]. DGAT2 inhibition blocks SREBP-1 cleavage and improves hepatic steatosis by increasing phosphatidylethanolamine in the ER. Cell Metabolism (Figure S7, panel D).
This protocol was developed based on previous research articles that used the budding assay in cultured cells:
Rexach et al. [2]. Distinct biochemical requirements for the budding, targeting, and fusion of ER-derived transport vesicles. The Journal of Cell Biology.
Rowe et al. [3]. COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. The Journal of Cell Biology.
Nohturfft et al. [1]. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell.
We also validated this protocol using transgenic rats that constitutively express HA-tagged human full-length (precursor) SREBP-1c under the control of the apoE promoter (TghSREBP-1c) [10] (Figure 3). The rats were either fasted for 24 h (fast) (low levels of SREBP-1 activation) or for 18 h and then re-fed a high carbohydrate diet for 6 h (re-fed) (high levels of SREBP-1 activation) before microsome/vesicle isolation to achieve the maximal differences in SREBP-1 activation. The isolated budding vesicles were validated by immunoblot with anti-calnexin and anti-ERGIC-53 antibodies. Calnexin, a resident ER protein, was not detectable in the budding vesicles (Figure 3, lanes 3 and 4), while ERGIC-53, a protein that migrates together with budding vesicles [11,12], was detected in both microsomal membranes and budding vesicles, indicating the successful isolation of ER membrane–derived budding vesicles without microsomal contamination. The experiment also showed that SREBP-1 is enriched in the vesicles budded from microsomal membranes of re-fed rats (Figure 3, lane 4 vs. lane 3), while ERGIC-53, serving as a loading control for budding vesicles, is similar between fast and re-fed groups, indicating that re-feeding the high carbohydrate diet increased SREBP-1 activation.
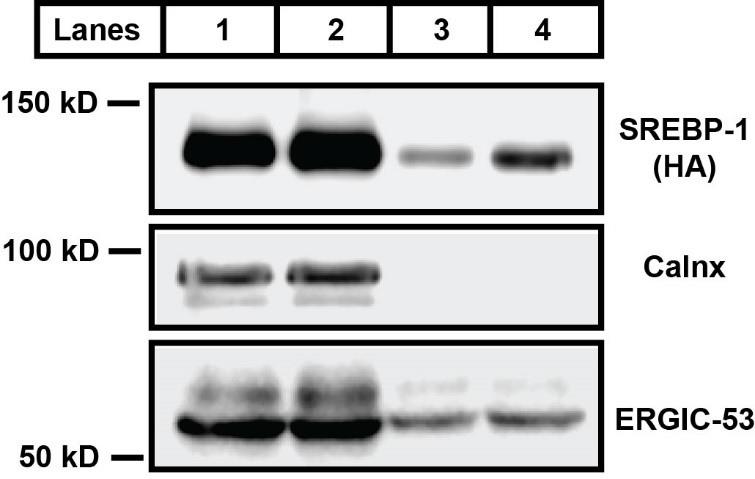
Figure 3. Immunoblot analysis of sterol regulatory element binding protein 1 (SREBP-1) in microsomal membranes and budding vesicles. TghSREBP-1c rats were fasted for 24 h (fast) or for 18 h and then re-fed a high carbohydrate diet for 6 h (re-fed). Liver microsomal membranes and budding vesicles were prepared for immunoblot analysis. Lane 1 represents microsomal membranes from fasted rats; lane 2 is microsomal membranes from re-fed rats; lane 3 is budding vesicles from fasted rat microsomes; lane 4 is budding vesicles from re-fed rat microsomes. The full length of transgenic human SREBP-1c protein (molecular weight ~ 125 kD) was detected using an anti-HA antibody. Calnexin (molecular weight ~ 95 kD) is a loading marker for microsome membranes. ERGIC-53 (molecular weight ~ 53 kD) is a protein associated with the vesicles during the budding process.Acknowledgments
We thank Dr. Jay D. Horton for critical reading of the manuscript. This work was supported by NIH grants 5P01HL160487 and 5P30DK127984. This protocol was developed based on previous research articles that used cultured cells for budding assay [1–3] and has been used and validated in Cell Metabolism (2024), DOI: 10.1016/j.cmet.2024.01.011 [8].
Competing interests
The authors have no competing interests to claim.
Ethical considerations
The animal experiments were performed with the approval of the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center.
References
- Nohturfft, A., Yabe, D., Goldstein, J. L., Brown, M. S. and Espenshade, P. J. (2000). Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 102(3): 315–323. https://doi.org/10.1016/s0092-8674(00)00037-4.
- Rexach, M. F. and Schekman, R. W. (1991). Distinct biochemical requirements for the budding, targeting, and fusion of ER-derived transport vesicles. J Cell Biol. 114(2): 219–229. https://doi.org/10.1083/jcb.114.2.219.
- Rowe, T., Aridor, M., McCaffery, J. M., Plutner, H., Nuoffer, C. and Balch, W. E. (1996). COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. J Cell Biol. 135(4): 895–911. https://doi.org/10.1083/jcb.135.4.895.
- Paulik, M., Nowack, D. D. and Morre, D. J. (1988). Isolation of a vesicular intermediate in the cell-free transfer of membrane from transitional elements of the endoplasmic reticulum to Golgi apparatus cisternae of rat liver. J Biol Chem. 263(33): 17738–17748.
- Schekman, R. and Orci, L. (1996). Coat proteins and vesicle budding. Science. 271(5255): 1526–1533. https://doi.org/10.1126/science.271.5255.1526.
- Shimomura, I., Shimano, H., Korn, B. S., Bashmakov, Y. and Horton, J. D. (1998). Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J Biol Chem. 273(52): 35299–35306. https://doi.org/10.1074/jbc.273.52.35299.
- Horton, J. D., Goldstein, J. L. and Brown, M. S. (2002). SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 109(9): 1125–1131. https://doi.org/10.1172/JCI15593.
- Rong, S., Xia, M., Vale, G., Wang, S., Kim, C. W., Li, S., McDonald, J. G., Radhakrishnan, A. and Horton, J. D. (2024). DGAT2 inhibition blocks SREBP-1 cleavage and improves hepatic steatosis by increasing phosphatidylethanolamine in the ER. Cell Metab. 36(3): 617–629 e617. https://doi.org/10.1016/j.cmet.2024.01.011.
- Brown, M. S. and Goldstein, J. L. (2009). Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL. J Lipid Res. 50 Suppl: S15–27. https://doi.org/10.1194/jlr.R800054-JLR200.
- Owen, J. L., Zhang, Y., Bae, S. H., Farooqi, M. S., Liang, G., Hammer, R. E., Goldstein, J. L. and Brown, M. S. (2012). Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc Natl Acad Sci USA. 109(40): 16184–16189. https://doi.org/10.1073/pnas.1213343109.
- Klumperman, J., Schweizer, A., Clausen, H., Tang, B. L., Hong, W., Oorschot, V. and Hauri, H. P. (1998). The recycling pathway of protein ERGIC-53 and dynamics of the ER-Golgi intermediate compartment. J Cell Sci. 111 (Pt 22): 3411–3425. https://doi.org/10.1242/jcs.111.22.3411.
- Appenzeller, C., Andersson, H., Kappeler, F. and Hauri, H. P. (1999). The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat Cell Biol. 1(6): 330–334. https://doi.org/10.1038/14020.
Article Information
Publication history
Received: Sep 18, 2024
Accepted: Oct 21, 2024
Available online: Nov 4, 2024
Published: Dec 20, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Xia, M., Edwards, T. and Rong, S. (2024). Assessment of SREBP Activation Using a Microsomal Vesicle Budding Assay. Bio-protocol 14(24): e5139. DOI: 10.21769/BioProtoc.5139.
Category
Molecular Biology > Protein > Activity
Cell Biology > Cell-based analysis > Protein maturation
Cell and Molecular Biology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









