- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
(*contributed equally to this work) Published: Vol 14, Iss 24, Dec 20, 2024 DOI: 10.21769/BioProtoc.5138 Views: 1789
Reviewed by: Ritu GuptaPrajita PandeySoumya Moonjely

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Separating Inner and Outer Membranes of Escherichia coli by EDTA-free Sucrose Gradient Centrifugation
Sheng Shu and Wei Mi
Mar 20, 2023 2915 Views

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1774 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1824 Views
Abstract
Cyclic diadenosine monophosphate (c-di-AMP) is a recently discovered second messenger that modulates several signal transduction pathways in bacterial and host cells. Besides the bacterial system, c-di-AMP signaling is also connected with the host cytoplasmic surveillance pathways (CSP) that induce type-I IFN responses through STING-mediated pathways. Additionally, c-di-AMP demonstrates potent adjuvant properties, particularly when administered alongside the Bacillus Calmette–Guérin (BCG) vaccine through mucosal routes. Because of its pivotal role in bacterial signaling and host immune response, this molecule has garnered significant interest from the pharmaceutical industry. This protocol outlines the quantification of c-di-AMP by an HPLC-based assay to enumerate the activity of c-di-AMP synthase from Mycobacterium smegmatis. The following protocol is designed to be generic, enabling the study of c-di-AMP synthase activity from other bacterial species. However, modifications may be required depending on the specific activity of c-di-AMP synthase from different bacterial sources.
Key features
• Easy and time-saving HPLC-based quantification method for c-di-AMP.
• Quick and reliable method to study enzymatic activity/kinetics of DisA/DAC.
• Elimination of potentially hazardous radioactive substrates and products for c-di-AMP quantification.
Keywords: C-di-AMPGraphical overview
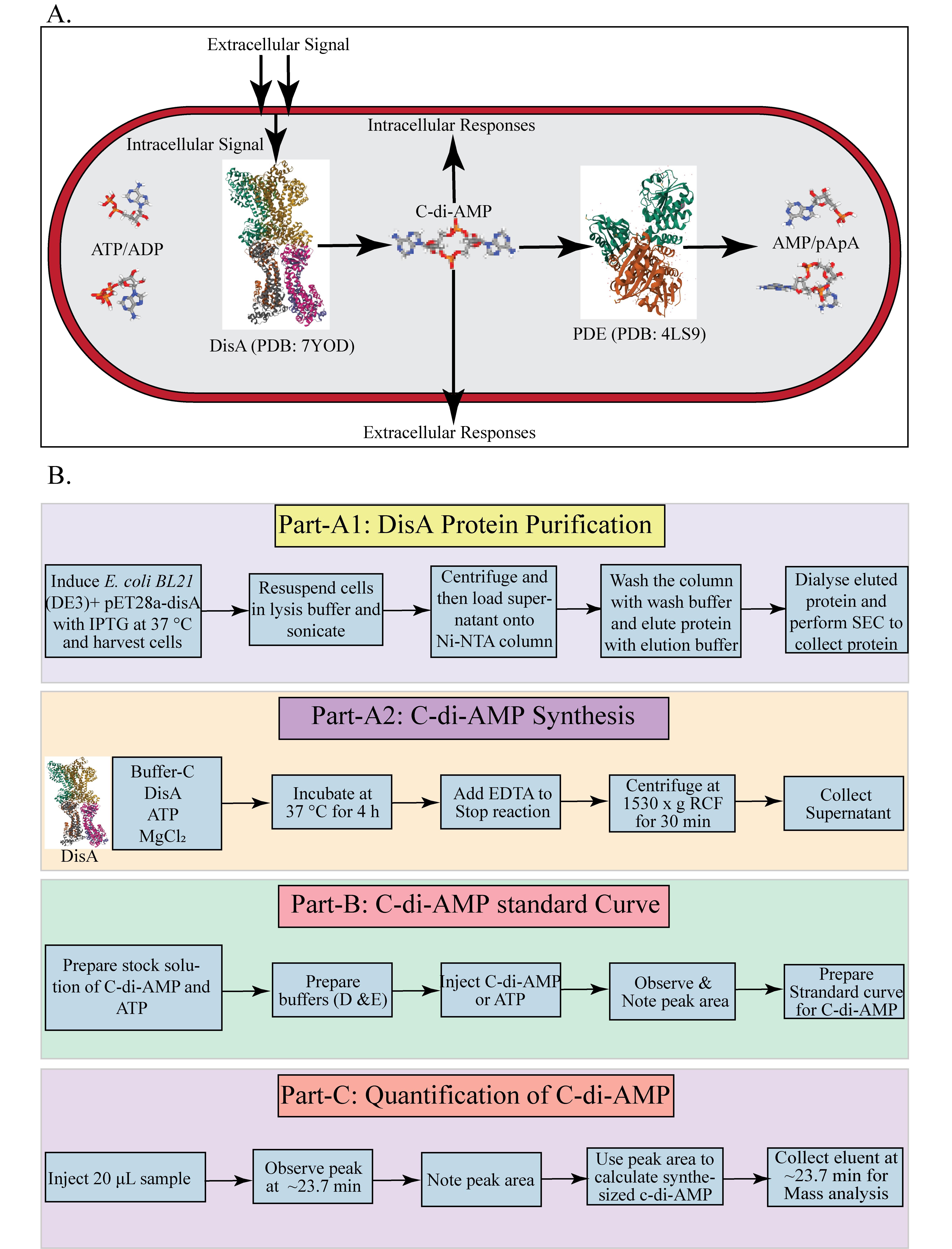
C-di-AMP homeostasis in mycobacteria and HPLC-based quantification process. A. Overview of c-di-AMP synthesis and hydrolysis in M. smegmatis. B. Overview of DisA protein purification, in vitro C-di-AMP synthesis, and HPLC-based quantification process.
Background
Cyclic di-adenosine monophosphate (c-di-AMP) is an essential secondary messenger that plays a pivotal role in regulating various physiological processes, including osmoregulation, DNA integrity maintenance, sporulation, cell wall homeostasis, ion-channel regulation, antibiotic resistance, virulence gene expression, acid resistance, and carbon metabolism [1,2]. In the bacterial kingdom, c-di-AMP synthesis is catalyzed by di-adenylate cyclase (DAC) domain–containing proteins, which convert two ATP molecules into c-di-AMP in the presence of metal ions such as Mg and Mn. To date, five classes of DAC domain–containing proteins have been identified in bacteria: DNA integrity scanning protein A (DisA), c-di-AMP synthase A (CdaA), c-di-AMP synthase sporulation-specific (CdaS), c-di-AMP synthase N-terminal TM segment (CdaM), and c-di-AMP synthase Z (CdaZ) [3,4]. DisA, an octameric protein, has dual functions: it can bind to Holliday junction DNA or synthesize c-di-AMP [1]. Mycobacterium smegmatis is a non-pathogenic, rapidly growing bacterium from the genus Mycobacterium. It serves as a widely used model organism for studying mycobacteria, including pathogenic species like Mycobacterium tuberculosis and non-tuberculous mycobacteria (NTM) pathogens, such as Mycobacterium abscessus and Mycobacterium avium [5]. In M. smegmatis, the gene MSMEG_6080 encodes the c-di-AMP synthase DisA, which catalyzes c-di-AMP synthesis in the presence of Mg ions. DisA contains three main domains: the N-terminal DAC domain-1, the C-terminal DNA binding domain-3, and a linker domain (domain-2) that connects domain 1 and domain 2 [4,6]. The enzyme's active site is located at the interface of the DAC domains [1]. The cellular concentration of c-di-AMP is regulated by phosphodiesterases (PDEs), which are located in different operons and contain DHH-DHHA1 or HD (His-Asp) domains [7–9]. PDEs hydrolyze c-di-AMP into pApA or AMP. These enzymes typically feature a unique structural alignment, consisting of an N-terminal linked to a degenerate GGDEF domain and a C-terminal DHH-DHHA1 module, essential for c-di-AMP hydrolysis [9]. In Mycobacterium tuberculosis, the PDE (CnpB) has a core DHH-DHHA1 domain that hydrolyzes c-di-AMP first to linear 5'-pApA and then to two 5'-AMP molecules [8]. Similarly, the PDE in M. smegmatis (MSMEG_2630) contains a DHH-DHHA1 domain without additional regulatory domains found in the GdpP protein family of Bacillus subtilis. All types of PDEs require specific divalent metal ions (Mg, Mn, Co) for their activity [1,2,7]. The opposing activities of these enzymes maintain the homeostasis of c-di-AMP within bacterial cells. Herein, we describe an HPLC-based protocol to study the c-di-AMP synthesis by DisA protein from M. smegmatis. This protocol outlines a straightforward method for quantifying c-di-AMP synthesized by mycobacterial DisA. The method is adapted from protocols by Bai et al., Christen et al., and Ryjenkov et al., with necessary modifications [10–13]. This HPLC-based method allows quick, easy, and quantitative estimation of synthesized c-di-AMP by DisA, avoiding the utilization of potentially hazardous radioactive substrates and products.
Materials and reagents
Reagents
Purified DisA protein (Origin: M. smegmatis)
ATP (Sigma, catalog number: A2383)
C-di-AMP (Jena Bioscience, catalog number: NU-954S)
Tris base (Sigma, catalog number: 10708976001)
EDTA (Sigma, catalog number: E4884)
Sodium chloride (NaCl) (Sigma, catalog number: S9888)
Tetrabutylammonium hydrogen sulfate (Sigma, catalog number: 15583)
Methanol (HPLC grade) (Sigma, catalog number: 34860)
Double-distilled water (Milli-Q Ultrapure Water Systems)
KH2PO4 (Sigma, catalog number: P5655)
MgCl2 (Sigma, catalog number: M8266)
Tris-Cl (Sigma, catalog number: 10812846001)
Luria Bertani Broth, Miller (HIMEDIA, catalog number: M1245)
IPTG (Sigma, catalog number I6758)
Dialysis membrane (Sigma, catalog number: D9527)
Pipette tips (Tarsons, catalog numbers: 521000, 521010, 521020)
1.5 mL reaction tubes (Tarsons, catalog number: 500010)
Membrane filter, 0.22 μm pore size (Merck, catalog number: GSWP04700)
Protein purification buffers: lysis buffer, wash buffer, and elution buffer (see Recipes)
DisA dialysis buffer (buffer A) (see Recipes)
Size exclusion chromatography (SEC) (buffer B) (see Recipes)
Buffer C for c-di-AMP synthesis reaction (see Recipes)
Buffer D (see Recipes)
Buffer E (see Recipes)
Protein Purification buffers
Lysis buffer
50 mM Tris-Cl (pH 7.9), 300 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride (PMSF)
Wash buffer
50 mM Tris-Cl (pH 7.9), 300 mM NaCl, and 40 mM imidazole
Elution buffer
50 mM Tris-Cl (pH 7.9), 300 mM NaCl, and 300 mM imidazole
DisA dialysis buffer (buffer A)
50 mM Tris-Cl buffer at pH 7.5, 300 mM NaCl
Size exclusion chromatography (SEC) (buffer B)
50 mM Tris-Cl (pH 7.9), 300 mM NaCl
Buffer C for c-di-AMP synthesis reaction
50 mM Tris (pH 9.4), 300 mM NaCl
Buffer D
100 mM KHPO, 4 mM tetrabutylammonium hydrogen sulfate, pH 5.9
Buffer E
75% (v/v) buffer D, 25% (v/v) methanol
Equipment
Micropipettes with varying capacity (T-2, T-10, T-20, T-100, T-200, T-1000) (Tarsons, catalog numbers: 030000, 030010, 030020, 030030, 030040, 03005)
Spectrophotometer (Eppendorf, model: BioSpectrometer®)
HPLC system: Agilent 1200 HPLC (Agilent Technologies, model: Agilent 1200 HPLC) equipped with quaternary pump, autosampler, thermostated column compartment, with a UV detector
Äkta (Cytiva, former GE Health care, model: 29707638)
C-18 column (4.6 × 150 mm) (Agilent, model: Eclipse XDB-C-18)
Superose 12 10/300 Column (Cytiva, catalog number: GE17-5173-01)
Dry bath (Bio-Rad, catalog number: 1660563)
Refrigerated centrifuge (Eppendorf, model: centrifuge 5430)
Vortex shaker (Tarsons, model: SPINIXTM)
Biological safety cabinet (Thermo Scientific, model: 1300 Series Class II, Type A2)
Vacuum pump and assembly (Tarsons, model: ROCKYVACTM Vacuum Pump with assembly)
Water purification system (Millipore, model: Ultra-Pure Water Purification System)
Software and datasets
HPLC system software LC/CE Agilent ChemStation (B.04.02 SP1, 4/2010)
Graph Pad Prism 5.01 (5.01, 9/2007)
Procedure
DisA protein purification and C-di-AMP synthesis assay with DisA protein from Mycobacterium smegmatis
Purify the DisA protein as described in Gautam et al. [12]. The protein purification procedure is briefly described below.
Inoculate E. coli BL21 (DE3) carrying the pET28a plasmid with the disA gene in LB medium and grow overnight at 37 °C. Prepare secondary cultures by inoculating 1% of the primary culture and grow at 37 °C with shaking until the OD reaches 0.6.
Induce the secondary cultures by adding 1 mM isopropyl ß-D-thiogalactopyranoside (IPTG) and incubate for 3 h at 37 °C.
Harvest the cultures by centrifugation at 495× g; then, resuspend the pellet in lysis buffer [50 mM Tris-Cl (pH 7.9), 300 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride (PMSF)].
Sonicate the cells and centrifuge at 1980× g to remove the cell debris.
Load the supernatant onto a Ni-NTA column to allow the protein to bind to the Ni-NTA beads.
Wash the column with 100 column volumes of wash buffer (50 mM Tris-Cl (pH 7.9), 300 mM NaCl, and 40 mM imidazole).
Elute the DisA protein using elution buffer containing 50 mM Tris-Cl (pH 7.9), 300 mM NaCl, and 300 mM imidazole. Analyze the protein by 10% SDS-PAGE.
Dialyze the purified DisA protein in Buffer A (see Recipes) for 12–16 h at 4 °C) as per the mentioned protocol [12].
Inject the dialyzed protein into a size exclusion chromatography (SEC) column to further purify the protein against buffer B (see Recipes).
Store the purified protein at -20 °C or -80 °C for future use.
Determine the concentration of the DisA protein (photometric measurement at 280 nm/BCA method).
Calculate the amount of DisA protein required to achieve a concentration of 1 μM in a 50 μL reaction.
In a 1.5 mL Eppendorf tube, set up a 50 μL reaction containing DisA protein (1 μM), 0.5 mM ATP, 5 mM MgCl in Buffer C (see Recipes).
Incubate the reaction at 37 °C for 4 h.
Stop the reaction by adding 10 mM EDTA to the reaction tube.
Centrifuge the reaction sample at 1,530× g for 30 min at 4 °C.
Collect the supernatant for HPLC analysis or store it at -20 °C for future use (Figure 1).
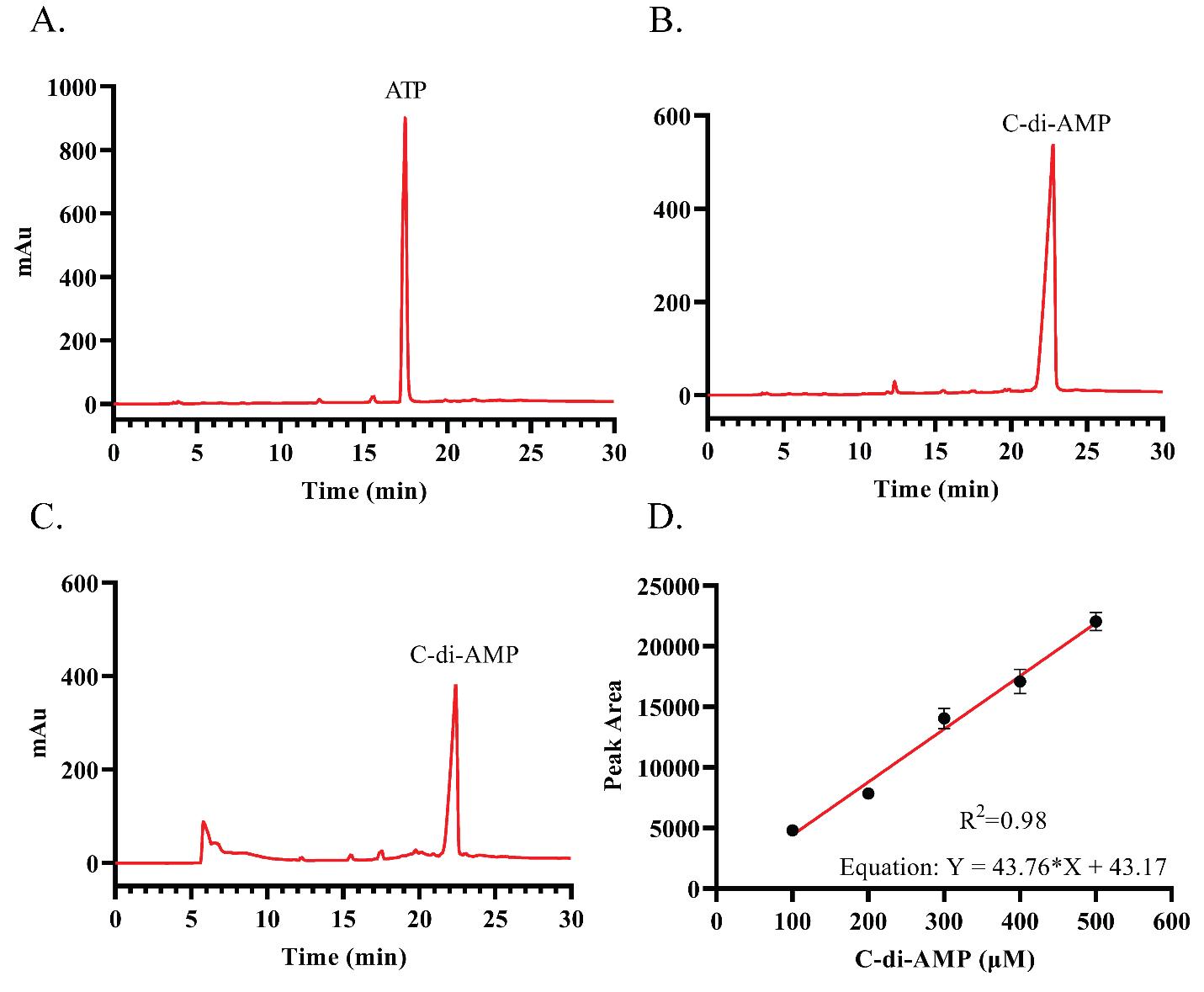
Figure 1. HPLC profiles of nucleotides and standard curve of c-di-AMP. A. HPLC profile of standard ATP. B. HPLC profile of standard C-di-AMP. C. HPLC profile of C-di-AMP synthesis reaction. D. Standard curve for c-di-AMP.
Generation of c-di-AMP standard curve
Prepare different concentrations of c-di-AMP or ATP (ranging from 100 to 500 μM) stock solutions in buffer C (see Recipes).
Use a reverse phase C-18 column to separate the nucleotides by using Buffer D (see Recipes) and Buffer E (see Recipes).
Filter and degas the HPLC buffers D and E using 0.22 μm filter paper.
Apply the following buffer gradient to separate the nucleotides at a flow rate of 0.7 mL/min:
0.0 min: 0% buffer E
2.5 min: 0% buffer E
5.0 min: 30% buffer E
10.0 min: 60% buffer E
14.0 min: 100% buffer E
21.0 min: 100% buffer E
22.0 min: 50% buffer E
23.0 min: 0% buffer E
Inject 20 μL of 500 μM pure c-di-AMP and then ATP separately for HPLC analysis.
After each sample injection, wash the HPLC column with buffer D for 10 min at a flow rate of 0.7 mL/min.
Pure c-di-AMP should show a peak at ~23.7 min, and ATP should show a peak at ~17.4 min.
After this preliminary validation, inject 20 μL of different concentrations (100–500 μM) of c-di-AMP into the HPLC and analyze using the previously stated gradient and buffers.
Use the chromatography data system software to obtain the peak area.
Prepare the standard curve by plotting the peak area under the curve (AUC) vs. the concentration of c-di-AMP used.
Analysis of the C-di-AMP synthesis assay reaction by HPLC
Use 20 μL of reaction sample for HPLC analysis.
Inject the reaction sample into the HPLC system using the gradient described above.
Observe for a peak at approximately ~23.7 min.
If a peak is observed at ~23.7 min, determine the area under the peak using the chromatography data system software.
Use the peak area to apply the standard curve and linear equation to determine the concentration (μM) of c-di-AMP formed in the reaction.
Collect the HPLC elution fractions around 23.7 min to further confirm the mass of the reaction product (See General note 2).
General notes and troubleshooting
The synthesis of c-di-AMP by DisA (a DAC domain-containing protein) highly depends on assay conditions like buffer compositions (NaCl/KCl), pH (5.4–9.4), temperature (35–40 °C), and incubation time (0–6 h). For Mycobacterium smegmatis, the highest activity was observed in Buffer C (50 mM Tris, 300 mM NaCl, pH 9.4). After 4 h of incubation at 37 °C, complete utilization of ATP was noted. When using DisA from different sources, activity optimization might be necessary to avoid multiple peaks (e.g., ATP or intermediate products).
We employ an earlier reported LC-MS method to analyze the HPLC eluted fractions from the DisA reactions (Burker Daltonics, Germany) as described by [14]. MS–MS analysis (negative or positive ion mode) of the eluted molecular masses is performed to verify the presence of c-di-AMP. The standard protocol involves LC–MS and MS–MS analysis using the HPLC gradient composition, as previously reported [15].
Validation of protocol
This protocol or parts of it has been used and validated in the following research article:
Ryjenkov DA et al. [10]. Cyclic diguanylate is a ubiquitous signalling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. [Fig 2B, Fig 3B,C, Used for c-di-GMP analysis].
Christen M et al. [11]. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem. [Fig 2A, Used for c-di-GMP analysis].
Bai Y et al. [13]. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS One. [Fig 2A, Used for C-di-AMP analysis]
Acknowledgments
D.C. gratefully acknowledges the initial financial help through a grant from the Dept. of Biotechnology, Govt. Of India, through a grant number DBT/PR33123/MED/29/1497/2020.
Competing interests
The authors declare no competing interests.
References
- Stülke, J. and Krüger, L. (2020). Cyclic di-AMP Signaling in Bacteria. Annu Rev Microbiol. 74(1): 159–179. https://doi.org/10.1146/annurev-micro-020518-115943
- Corrigan, R. M. and Gründling, A. (2013). Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 11(8): 513–524. https://doi.org/10.1038/nrmicro3069
- Zhang, L. and He, Z. G. (2013). Radiation-sensitive Gene A (RadA) Targets DisA, DNA Integrity Scanning Protein A, to Negatively Affect Cyclic Di-AMP Synthesis Activity in Mycobacterium smegmatis. J Biol Chem. 288(31): 22426–22436. https://doi.org/10.1074/jbc.m113.464883
- Petchiappan, A., Mahapa, A. and Chatterji, D. (2020). Cyclic Dinucleotide Signaling in Mycobacteria. In: Chou, SH., Guiliani, N., Lee, V., Römling, U. (eds) Microbial Cyclic Di-Nucleotide Signaling. Springer, Cham. 3–25. https://doi.org/10.1007/978-3-030-33308-9_1
- Sparks, I. L., Derbyshire, K. M., Jacobs, W. R. and Morita, Y. S. (2023). Mycobacterium smegmatis: The Vanguard of Mycobacterial Research. J Bacteriol. 205(1): e00337–22. https://doi.org/10.1128/jb.00337-22
- Witte, G., Hartung, S., Büttner, K. and Hopfner, K. P. (2008). Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates. Mol Cell. 30(2): 167–178. https://doi.org/10.1016/j.molcel.2008.02.020
- Tang, Q., Luo, Y., Zheng, C., Yin, K., Ali, M. K., Li, X. and He, J. (2015). Functional Analysis of a c-di-AMP-specific Phosphodiesterase MsPDE from Mycobacterium smegmatis. Int J Biol Sci. 11(7): 813–824. https://doi.org/10.7150/ijbs.11797
- He, Q., Wang, F., Liu, S., Zhu, D., Cong, H., Gao, F., Li, B., Wang, H., Lin, Z., Liao, J., et al. (2016). Structural and Biochemical Insight into the Mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP Phosphodiesterase. J Biol Chem. 291(7): 3668–3681. https://doi.org/10.1074/jbc.m115.699801
- Commichau, F. M., Heidemann, J. L., Ficner, R. and Stülke, J. (2019). Making and Breaking of an Essential Poison: the Cyclases and Phosphodiesterases That Produce and Degrade the Essential Second Messenger Cyclic di-AMP in Bacteria. J Bacteriol. 201(1): e00462–18. https://doi.org/10.1128/jb.00462-18
- Ryjenkov, D. A., Tarutina, M., Moskvin, O. V. and Gomelsky, M. (2005). Cyclic Diguanylate Is a Ubiquitous Signaling Molecule in Bacteria: Insights into Biochemistry of the GGDEF Protein Domain. J Bacteriol. 187(5): 1792–1798. https://doi.org/10.1128/jb.187.5.1792-1798.2005
- Christen, M., Christen, B., Folcher, M., Schauerte, A. and Jenal, U. (2005). Identification and Characterization of a Cyclic di-GMP-specific Phosphodiesterase and Its Allosteric Control by GTP. J Biol Chem. 280(35): 30829–30837. https://doi.org/10.1074/jbc.m504429200
- Gautam, S., Mahapa, A., Yeramala, L., Gandhi, A., Krishnan, S., Kutti R., V. and Chatterji, D. (2023). Regulatory mechanisms of c‐di‐AMP synthase from Mycobacterium smegmatis revealed by a structure: Function analysis. Protein Sci. 32(3): e4568. https://doi.org/10.1002/pro.4568
- Bai, Y., Yang, J., Zhou, X., Ding, X., Eisele, L. E. and Bai, G. (2012). Mycobacterium tuberculosis Rv3586 (DacA) Is a Diadenylate Cyclase That Converts ATP or ADP into c-di-AMP. PLoS One. 7(4): e35206. https://doi.org/10.1371/journal.pone.0035206
- Bharati, B. K., Mukherjee, R. and Chatterji, D. (2018). Substrate-induced domain movement in a bifunctional protein, DcpA, regulates cyclic di-GMP turnover: Functional implications of a highly conserved motif. J Biol Chem. 293(36): 14065–14079. https://doi.org/10.1074/jbc.ra118.003917
- Bharati, B. K., Sharma, I. M., Kasetty, S., Kumar, M., Mukherjee, R. and Chatterji, D. (2012). A full-length bifunctional protein involved in c-di-GMP turnover is required for long-term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology (N Y). 158(6): 1415–1427. https://doi.org/10.1099/mic.0.053892-0
Article Information
Publication history
Received: Jul 30, 2024
Accepted: Oct 4, 2024
Available online: Nov 4, 2024
Published: Dec 20, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Mahapa, A., Gautam, S., Rathore, A. and Chatterji, D. (2024). An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis. Bio-protocol 14(24): e5138. DOI: 10.21769/BioProtoc.5138.
Category
Microbiology > Microbial biochemistry > Other compound
Biochemistry > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









