- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Novel Cross-Species Salivary Gland-Parasympathetic Neuron Coculture System
(*contributed equally to this work) Published: Vol 14, Iss 21, Nov 5, 2024 DOI: 10.21769/BioProtoc.5101 Views: 1964
Reviewed by: Xiaokang WuKrishna Murthy NakuluriAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Human iPSC-Derived Neuron and Oligodendrocyte Co-culture as a Small-Molecule Screening Assay for Myelination
Stefanie Elke Chie [...] Maria Consolata Miletta
May 5, 2025 3362 Views
Spheroid Sheets: A Scalable Platform for Producing Tissue Membrane Constructs
Quang Bach Le [...] Deepak Choudhury
Nov 20, 2025 1511 Views
Vascularization of Human Pancreatic Islets With Adaptive Endothelial Cells for In Vitro Analysis and In Vivo Transplantation
Ge Li [...] Shahin Rafii
Dec 20, 2025 720 Views
Abstract
The parasympathetic nervous system is essential for salivary gland development and functionality. Parasympathetic neuron (parasymN) innervation is the main neural network that controls salivary secretion. Therefore, an exclusive model to study parasympathetic neurons and salivary gland tissue circuitry will significantly improve the understanding of the role of parasymN activation on salivary regulation. Harvesting primary rodent parasymNs is challenging due to their body-wide disbursed location. Similarly, the salivary glands are distributed in various locations around and within the oral cavity. Here, we present a coculture model system using human pluripotent stem cell (hPSC)-derived parasymNs and primary mouse von Ebner’s gland cells. We previously reported the first protocol to robustly generate human parasymNs from hPSCs through the Schwann cell precursor (SCP) lineage. The hPSC-parasymNs are functional and have been applied to model several autonomic disorders. We also used a Sox10-Cre::tdTomato (hereafter referred to as RFP) reporter mouse line, which labeled von Ebner’s glands, a type of minor salivary gland connected to the trough of circumvallate and foliate taste papillae. This labeling allowed for visualization and efficient isolation of primary tissues in young adult mice (8–10 weeks). By coculturing the two tissues, human parasymNs control mouse salivary gland cell growth and activation. Both parasymNs and primary salivary gland cells can be frozen and stocked at early stages of differentiation and isolation, making applications easier. This novel coculture model system could also be used to model and study related human diseases in the future, such as dry mouth syndrome.
Key features
• Differentiation of human parasymNs from hPSCs.
• Dissecting RFP+ mouse Ebner’s gland tissues for primary culture.
• Protocol to coculture human parasymNs with mouse primary salivary gland cells.
• Allows developmental and functional assessments of salivary regulation by the parasympathetic nervous system.
Keywords: Parasympathetic nervous systemGraphical overview
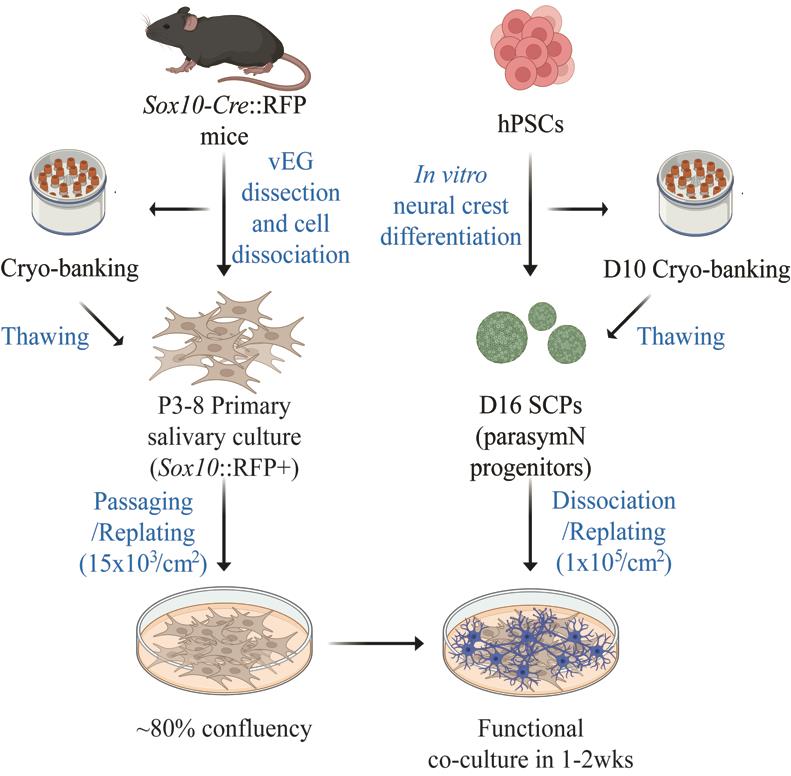
Background
The autonomic nervous system (ANS) regulates involuntary biological functions, such as blood pressure, heartbeat, and gland secretion. The ANS is subdivided into the sympathetic and parasympathetic nervous systems (SNS and PSNS). While sympathetic neurons (symNs) trigger the fight-or-flight response that increases the heartbeat, parasymNs trigger the rest-and-digest response to calm down the body, decreasing the heartbeat [1]. Clinically, the SNS usually draws more attention in autonomic dysregulation, because its dysfunction typically drives more dramatic and detrimental responses and symptoms [2,3]. However, innervation by parasymNs drives important functions, including salivation of the salivary glands [4]. Accordingly, PSNS activation induces elevated calcium (Ca2+) flux in salivary gland cells, which leads to increased saliva production [5].
Harvesting large numbers of primary parasymNs from rodents and humans for multiple assays or high-throughput screening is challenging. The hPSC technology can bridge this problem. hPSC-derived cells provide an unlimited source of any human cell type as desired. This is, however, highly dependent on good in vitro differentiation protocols. To date, there are several well-characterized symN differentiation protocols, including ours [6–11]. In contrast, only two parasymN protocols have been reported so far, and only our protocol generates functional hPSC-parasymNs that are derived via the proper developmental process, i.e., differentiation from neural crest cell (NCC)-derived SCPs [12]. In this protocol, the NCC fate was first induced from hPSCs. This was followed by SCP differentiation in 3D spheroid culture. Finally, SCP spheroids were replated to generate parasymNs [12]. To make research applications easier and secure reproducibility and efficiency, we showed that the protocol can be reproduced using frozen NCC stocks [12]. hPSC-parasymNs express specific genetic markers, including early autonomic markers ASCL1/PHOX2B, later cholinergic markers CHAT/VACHT/CHT/CHRM2/CHRM4, and specific parasymN markers HMX2/3[12,13]. We confirmed the electrical activity of hPSC-parasymNs, and electric activity could be manipulated via nicotine, which mimics preganglionic signaling. In cocultures with cardiomyocytes, nicotine activation downregulated their beating rate [12]. Using hPSC-parasymNs, we studied ANS responsiveness to COVID-19 infection and modeled the genetic autonomic disorder familial dysautonomia [12].
Salivary secretion, important for wetting oral surfaces and maintaining taste acuity, is dependent on the activity of innervating nerves to the salivary glands. Salivary glands include three pairs of major (parotid, submandibular, and sublingual) and numerous minor salivary glands around the mouth cavity. The von Ebner’s glands are a type of minor salivary glands with the ducts open to the trench of circumvallate and foliate taste papillae. Their connection to the taste papillae makes it easy to recognize and dissect. They are innervated by both sympathetic and parasympathetic efferent nerves, yet the salivary fluid secretion (water mobilizing) is largely controlled by parasympathetic activities [14].
Here, we describe a protocol to coculture hPSC-parasymNs with RFP+ mouse primary von Ebner’s gland cells [15,16]. We show that the proliferation of von Ebner’s gland cells is promoted when parasymNs are present. Furthermore, the number of auto-fluorescent secretory granule-positive von Ebner’s gland cells is significantly increased in the cocultures, suggesting the increased salivary cell maturation due to parasymN innervation. We also measured Ca2+ flux to represent salivary production, which is increased after parasymN stimulation in the cocultures. Impaired salivation can be seen in multiple diseases, such as Parkinson’s disease, Wilson disease, Angelman syndrome, various infections, and drug abuse [4]. Our newly established model may therefore be used to study the defects and cytotoxicity in those conditions, as well as the regenerative potential of hPSC-parasymNs on damaged salivary glands in the future.
Materials and reagents
Biological materials
Human embryonic stem cell line (WiCell, WA09, female, NIH 0062)
Sox10-Cre mice (Jackson Laboratory, stock 025807)
RFP Cre reporter mice B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (Jackson Laboratory, stock 007914)
Reagents
Geltrex (Invitrogen, catalog number: A1413202)
Poly-L-ornithine hydrobromide (PO) (Sigma, catalog number: P3655)
Mouse laminin I (LM) (R&D Systems, catalog number: 3400-010-01)
Human fibronectin (FN) (VWR/Corning, catalog number: 47743-654)
Essential 8 medium (E8) (Gibco, catalog number: A15169-01)
Essential 8 supplement (E8) (Gibco, catalog number: A15171-01)
Essential 6 medium (E6) (Gibco, catalog number: A15165-01)
Neurobasal media (Gibco, catalog number: 21103-049)
Stem cell banker (Amsbio, catalog number: 11924)
DMEM/F12 (Thermo Fisher/Life Technologies, catalog number: 11330-057)
DMSO (Thermo Fisher/Life Technologies, catalog number: BP231-100)
Fetal bovine serum (FBS) (Atlanta Biologicals, catalog number: S11150)
B27 supplement (Thermo Fisher/Life Technologies, catalog number: 12587-010)
N2 supplement (Thermo Fisher/Life Technologies, catalog number: 17502-048)
L-Glutamine (Thermo Fisher/Gibco, catalog number: 25030-081)
GlutaMAX (Gibco, catalog number: 35050061)
Antibiotic-antimycotic (Fisher, catalog number: 15 240 096)
Accutase (Innovation Cell Technologies, catalog number: AT104500)
EDTA (Sigma, catalog number: ED2SS)
Trypsin-EDTA (Gibco, catalog number: 25200056)
Phosphate-buffered saline (PBS) (Gibco, catalog number: 14190-136)
SB431542 (Tocris/R&D Systems, catalog number:1614), make 10 mM stock
BMP4 (R&D Systems, catalog number: 314-BP), make 10 µg/mL stock
CHIR99021 (R&D Systems, catalog number: 4423), make 6 mM stock
Y27632 (R&D Systems, catalog number: 1254), make 10 mM stock
FGF2 (R&D Systems, catalog number: 233-FB/CF), make 10 µg/mL stock
NRG1 (PeproTech, catalog number: 100-03), make 100 µg/mL stock
GDNF (PeproTech, catalog number: 450), make 10 µg/mL stock
BDNF (R&D Systems, catalog number: 248-BD), make 10 µg/mL stock
CNTF (R&D Systems, catalog number: 257-NT), make 100 µg/mL stock
Ascorbic acid (Sigma, catalog number: A8960), make 100 mM stock
dbcAMP (Sigma, catalog number: D0627), make 100 mM stock
Retinoic acid (Sigma, catalog number: R2625), make 1 mM stock
Collagenase A (Tribioscience, catalog number: TBS2116-01), make 2 mg/mL stock
Dispase II (Tribioscience, catalog number: TBS2117-01), make 5 mg/mL stock
DPBS without Ca+2 and Mg+2 (Thermo Fisher/Life Technologies, catalog number: 14190144)
70% ethanol
Solutions
NCC induction medium, D0, 1 (see Recipes)
NCC induction medium, D2-10 (see Recipes)
SCP differentiation medium, D10-16 (see Recipes)
ParasymN differentiation medium, D16 (see Recipes)
Salivary gland cell medium (see Recipes)
Tongue tissue separation solution (see Recipes)
Salivary gland cell dissociation solution (see Recipes)
Dissociation termination solution (see Recipes)
Recipes
NCC induction medium, D0, 1 (50 mL)
Reagent Final concentration Quantity or Volume E6 n/a 50 mL SB431542 (10 mM stock) 10 µM 50 µL BMP4 (10 µg/mL stock) 0.2–1 ng/mL 1–5 µL CHIR99021 (6 mM stock) 300 nM 2.5 µL Y27632 (10 mM stock) 10 µM 50 µL Note: Please see Troubleshooting 1 for more information.
NCC induction medium, D2-10 (100 mL)
Reagent Final concentration Quantity or Volume E6 n/a 100 mL SB431542 10 µM 100 µL CHIR99021 0.75 µM 12.53 µL SCP differentiation medium, D10-16 (100 mL)
Reagent Final concentration Quantity or Volume Neurobasal media n/a 100 mL N2 supplement 1% 1 mL B27 supplement 2% 2 mL L-glutamine 1% 1 mL CHIR99021 3 µM 50 µL FGF2 (10 µg/mL stock) 10 ng/mL 100 µL NRG1 (100 µg/mL stock) 10 ng/mL 10 µL ParasymN differentiation medium, D16 (100 mL)
Reagent Final concentration Quantity or Volume Neurobasal media n/a 100 mL N2 supplement 1% 1 mL B27 supplement 2% 2 mL GlutaMAX 1% 1 mL FBS 1% 1 mL GDNF (10 µg/mL stock) 25 ng/mL 250 µL BDNF (10 µg/mL stock) 25 ng/mL 250 µL Ascorbic acid (100 mM stock) 200 µM 200 µL CNTF (100 µg/mL stock) 25 ng/mL 10 µL dbcAMP (100 mM stock) 200 µM 200 µL Retinoic acid (1 mM stock) 0.125 µM 12.5 µL Note: Retinoic acid should be added to the medium freshly every feeding.
Salivary gland cell medium (10 mL)
Reagent Final concentration Quantity or Volume DMEM/F12 1× 9.6 mL FBS 1% 100 µL B27 1× 200 µL Antibiotic-antimycotic 1× 100 µL Tongue tissue separation solution (2 mL)
Reagent Final concentration Quantity or Volume Collagenase A (2 mg/mL stock) 1 mg/mL 1.0 mL Dispase II (5 mg/mL stock) 2.5 mg/mL 1.0 mL PBS 1× Salivary gland cell dissociation solution (3 mL)
Reagent Final concentration Quantity or Volume Trypsin-EDTA 0.25% 3 mL Dissociation termination solution (2 mL)
Reagent Final concentration Quantity or Volume DMEM/F12 1× 1800 µL FBS 10% 200 µL
Laboratory supplies
TC-treated 6-well tissue culture plate (Corning, catalog number: 3516)
TC-treated 24-well tissue culture plate (Corning, catalog number: 3526)
TC-treated 10 cm tissue culture plate (Corning, catalog number: 430167)
Ultra-low attachment plate (Corning, catalog numbers: 07 200 601 and 07 200 602)
15 mL conical tissue culture tubes (VWR/Corning, catalog number: 89039-664)
50 mL conical tissue culture tubes (VWR/Corning, catalog number: 89039-656)
Cryovial (Thermo Fisher/Life Technologies, catalog number: 375353)
Trypan blue (Corning, catalog number: MT-25-900-CI)
P2-10, 20, 200, 1000 pipettes and tips (Eppendorf)
Parafilm (ULINE)
5 mL low retention vials (Eppendorf, catalog number: 30122348)
35 mm dish (Genesee Scientific, catalog number: 32-103G)
100 mm culture dishes (Genesee Scientific, catalog number: 32-107G)
3 mL syringes (BD, catalog number: 8194938)
30-G needle (BD, catalog number: 9193532)
70 µm cell strainer (Fisher Scientific, catalog number: 352350)
35 µm cell strainer (Electron Microscopy Science, catalog number: 64750-25)
100% CO2 gas (Airgas, CD USP50)
Plastic CO2 euthanasia chamber (Length: 10 cm, wide: 7 cm, and height: 5 cm)
Equipment
Water bath (VWR)
CellDrop automated cell counter (DeNovix)
Racks
Liquid nitrogen tank (Custom Biogenic Systems)
Freezer (-20 °C)
Refrigerator (2–8 °C)
Water bath (37 °C)
Centrifuge (Eppendorf)
37 °C incubator with 5% CO2
Class II biological safety hood
Lionheart FX microscope (BioTek)
Dissecting microscope (Olympus, model: SZX16 upright fluorescent and brightfield)
Biosafety cabinet (NUAIRE, model: GellGard, Class II, Type A2)
CO2 incubator (NUAIRE)
Centrifuge (Thermo Scientific, model: LEGEND XTR)
Light microscope (EVOS XL Core)
Tank with CO2 (Airgas)
Instruments
Surgical and fine scissors (Moria, catalog number: 9601; Fine Science Tool, catalog number: 91604-09)
Surgical and fine forceps (Inox electronic, catalog number: 91150-20; Fine Science Tool, catalog number: 112900)
Spatula (Fine Science Tool, catalog number: 10360-13)
Software and datasets
Gen5 (BioTek)
Prism v10.0.3 (GraphPad, 9/25/2023)
Fiji (Image J)
Adobe Photoshop
CellSens Dimension
Procedure
Caution: The procedures for sections A and B will take 1.5–2.0 h without an opportunity to pause. The experiments need to be done as efficiently as possible for harvesting healthy cells. Please make sure you have all the instruments, solutions, and equipment ready before euthanizing the animals.
Salivary gland dissection
Euthanize the 8-week-old Sox10-Cre::RFP mice by placing them in a CO2 chamber. Ensure mouse death by cervical dislocation.
Place the mouse in the surgical area. Wet the mouse head using 70% ethanol to disinfect the surface and prevent fur from getting into the oral cavity.
Decapitate the mouse. Hold the head in one hand and open the mouth cavity by pulling the skin over the skull and below the mandible (Figure 1A).
Cut the corners of the mouth along the cheek using dissecting scissors to further open the oral cavity (Figure 1B).
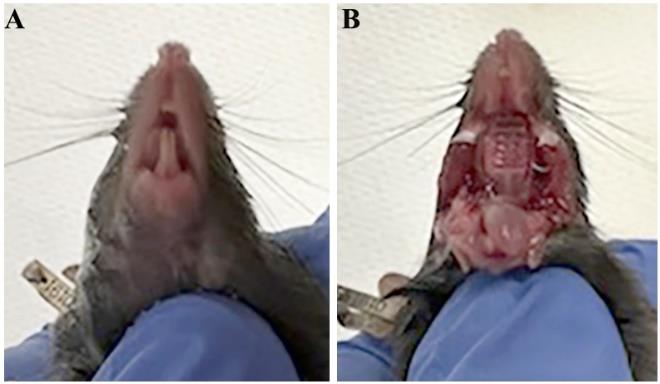
Figure 1. Opening the mouse oral cavity. (A) Image of ventral view of a mouse head. (B) Image of a mouse head with the mouth cavity widely open.Dissect the tongue with the mandible and remove the skin covering the mandible (Figure 2). Transfer the tongue on the mandible to 30 mL of fresh sterile DPBS (Ca+2 and Mg+2 free) in a 50 mL tube. Wash five times using DPBS.
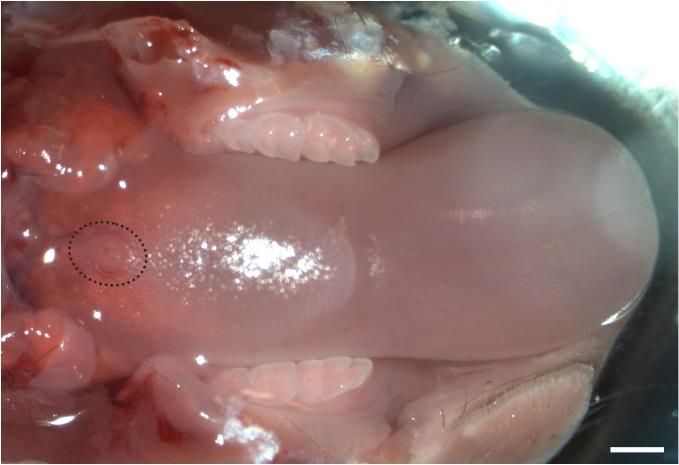
Figure 2. Dissected tongue with the mandible. A representative image of the dorsal view of an adult mouse tongue sitting on the mandible. Black dotted line encircles the single circumvallate taste papilla in the midline of posterior tongue. Scale bar: 500 μm.Using surgical forceps to hold the tongue under a dissecting microscope, inject ~0.5 mL of the tongue tissue separation solution (a mixture of Collagenase A and Dispase II enzymes) into the subepithelial space of posterior tongue as described [17] (Figure 3A and B) and incubate for 30 min at 37 °C (Figure 3C).
Note: See Troubleshooting #3 and #5 for precautions.
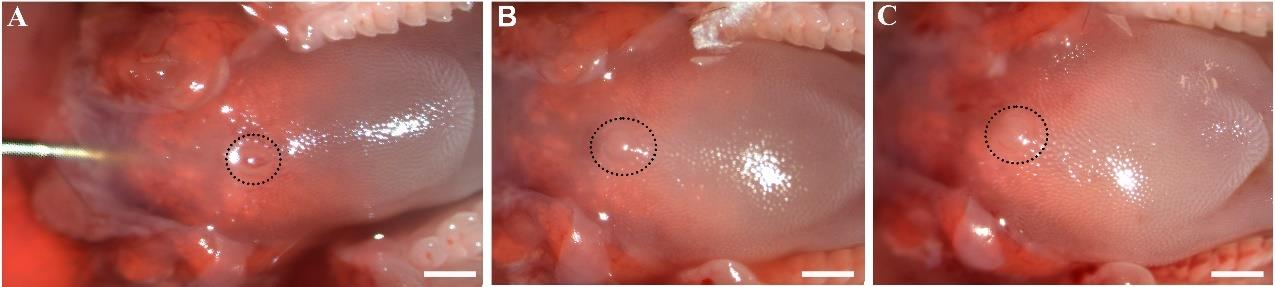
Figure 3. Intralingual injection of tongue tissue separation solution. (A) An image to illustrate the entry and location of the needle for injecting the solution into the posterior part of the tongue. (B) An image of dorsal view of the posterior tongue after 0.5 mL solution injection. (C) An image of dorsal view of the posterior tongue after 15 min of incubation at 37 °C. Black dotted lines encircle the single circumvallate taste papilla. Scale bars: 500 μm.After the incubation, dissect the posterior tongue region with RFP+ von Ebner’s glands. Use small scissors to cut the epithelium of the circumvallate papilla region (Figure 4A), then use fine forceps to peel off the epithelium of the circumvallate papilla region (Figure 4B).
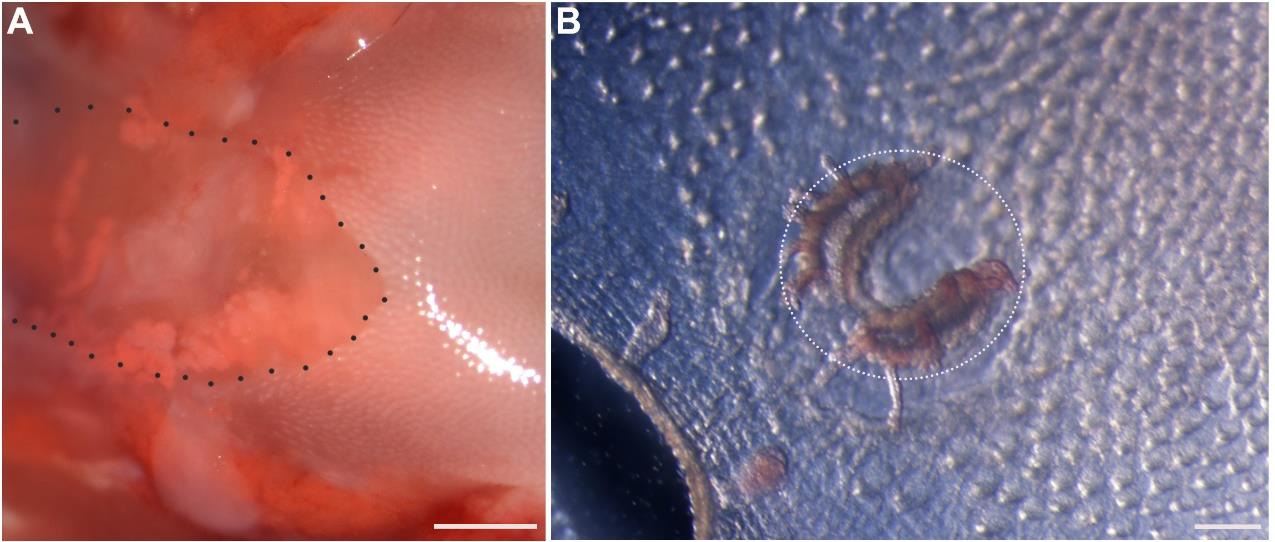
Figure 4. Peeling the circumvallate epithelium. (A) Brightfield image of the dorsal view of the posterior tongue region after removing part of the epithelium. Black dotted line outlines the cutting edge of epithelium in the circumvallate papilla area with the epithelium removed. (B) Brightfield image of the ventral view of the peeled epithelium. White dotted line encircles the circumvallate papilla epithelium. Scale bars: 500 μm in A; 100 μm in B.Under a fluorescent stereomicroscope, we were able to see the RFP+ von Ebner’s glands in the circumvallate region (Figure 5A). Dissect and collect ~0.1 mL of von Ebner’s salivary gland tissue in the surrounding of the circumvallate papilla under the fluorescent microscope (Figure 5B).
Caution: If you collect more gland tissues for more cells, you will need to use more solution of 0.25% trypsin-EDTA accordingly.
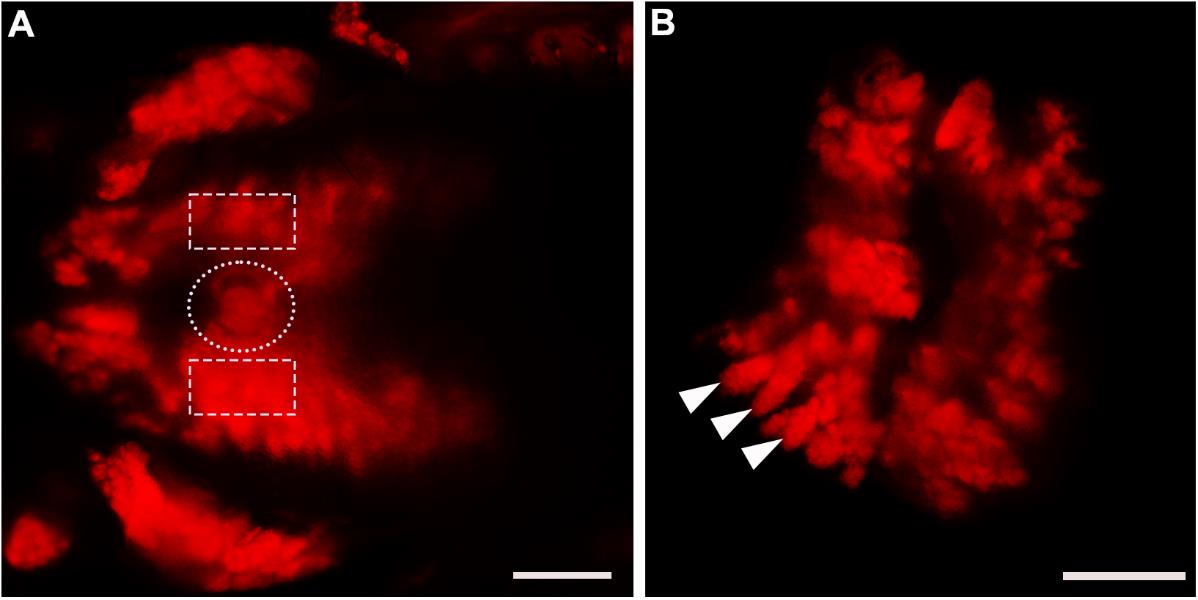
Figure 5. Dissection of von Ebner’s gland tissue from Sox10-Cre::RFP mice. (A) RFP fluorescent image of the dorsal view of the posterior tongue region under the fluorescent stereomicroscope. Bright RFP+ glands were readily visible in the regions surrounding and under the circumvallate papilla. White dotted line in A encircles the circumvallate papilla. White dashed squares encircle the circumvallate papilla-adjacent regions from which glands were collected. (B) RFP fluorescent image of two pieces of dissected tissue enriched RFP+ Ebner’s glands (white arrowheads). Scale bars: 500 μm.Wash the salivary glands in 15 mL of fresh sterile DPBS in a 100 mm sterile culture dish.
Salivary gland cell dissociation
Transfer salivary glands into a 35 mm dish.
Use fine-tip scissors to cut the salivary gland tissues into small pieces (Figure 6).
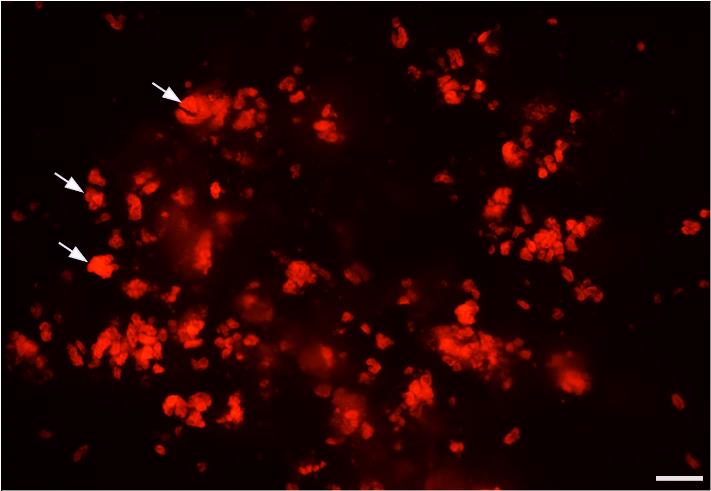
Figure 6. Tissue pieces of Ebner’s glands after mincing. RFP+ Ebner’s gland tissue (Figure 5B) was cut into smaller pieces using fine scissors under a fluorescent stereomicroscope. Arrows point to a few pieces of RFP+ Ebner’s gland tissue after mincing. Scale bar: 200 μm.Incubate the tissue pieces in 3 mL of salivary gland cell dissociation solution (0.25% trypsin-EDTA) for 30 min at 37 °C. Gently agitate the tissues every 5 min using 1 mL pipette tips. Do not cut the tip, as the sharp edge of the cutting end may injure the cells.
Note: See Troubleshooting 4 for precautions.
Add 1 mL of dissociation termination solution to stop the reaction and transfer all of the solution with cells into a 5 mL low-binding vial.
Centrifuge the cell suspension at 450× g for 10 min at 4 °C and remove the supernatant as much as possible. Be careful not to disturb the cell pellet.
To completely remove the remaining trypsin-EDTA, repeat step B4, adding 1 mL of dissociation termination solution to suspend the cells. Then, repeat step B5 for the centrifugation.
Remove the supernatant and leave one-fourth of the medium (250–300 mL) without disturbing the pellet and filter the cells using a 70 µm cell strainer, followed by a 35 µm cell strainer.
Collect the filtered cell suspension and proceed straight to Section C (Figure 7).
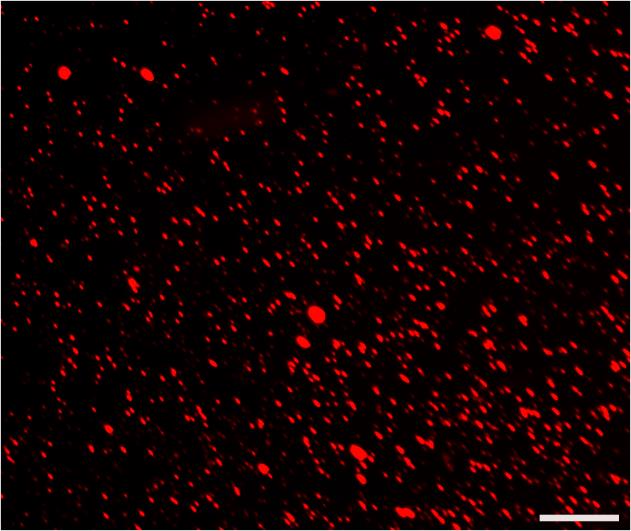
Figure 7. Cell suspension. Photomicrographs of the dissociated cells and some small cell clusters from RFP+ Ebner’s glands. Scale bar: 100 μm.
Salivary cell passaging
Centrifuge the cell suspension at 200× g for 5 min.
Aspirate the medium.
Wash cells twice with PBS (1 mL/24-well, 3 mL/6-well, and 4 mL/10 cm plates).
Add prewarmed (5 min in a water bath) trypsin-EDTA (1 mL/24-well, and 3 mL/10 cm plates).
Incubate for 5 min in a 37 °C incubator.
Pipette gently with a P1000 pipette 3–5 times to fully dissociate the cells.
Transfer the trypsin-EDTA and cell solution to a conical tissue culture tube that contains at least 5 times higher volume of salivary gland cell medium.
Centrifuge at 200× g for 5 min.
Resuspend the cells with 1 mL of salivary gland cell medium.
Count the cells.
Replate the cells on desired tissue culture plates at 15 × 103/cm2 density with salivary gland cell medium (0.5 mL/24-well, 2 mL/6-well, and 7 mL/10 cm plates).
Change the medium the next day, and every 2–3 days after (1 mL/24-well, 3 mL/6-well, and 10 mL/10 cm plates).
Cells should reach >80% confluency in one week (Figure 8) and will be ready for replating/freezing.
The ideal passage number of the cells for the experiment is P3–8. Please see also troubleshooting 2.
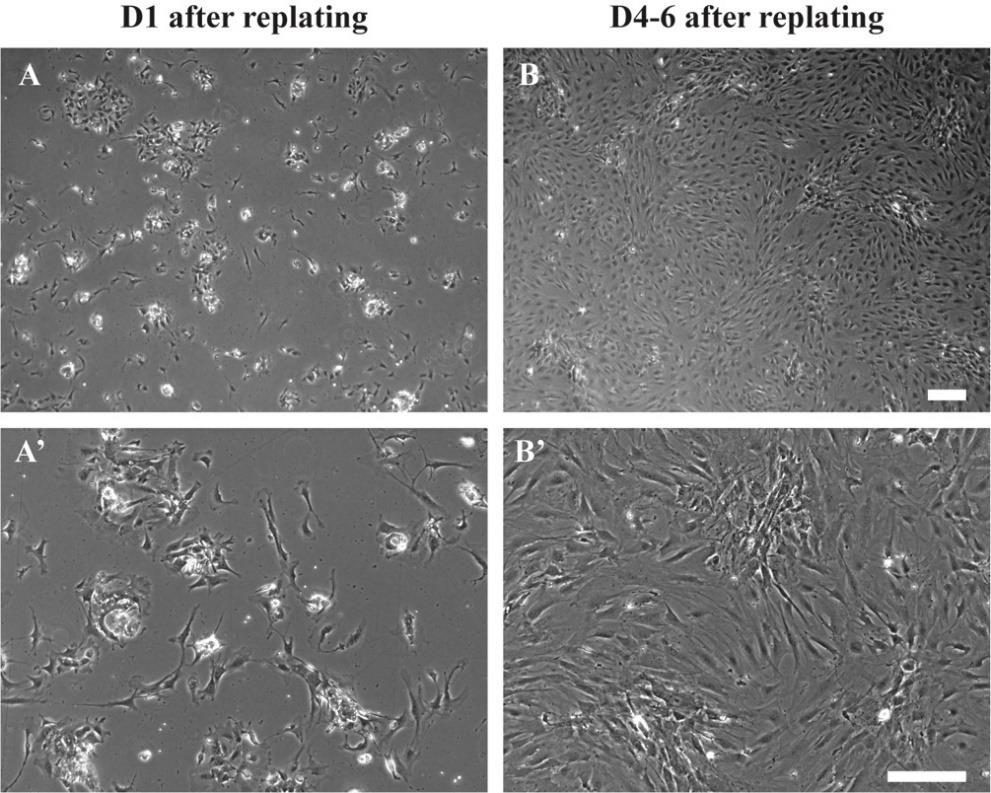
Figure 8. Cell morphology of salivary gland cells after replating. Brightfield images showing the isolated salivary gland cells after (A and A’) 24 h, and (B and B’) 4–6 days. Cell density in B is ideal for making frozen stocks or replating the SCPs to start the coculture. Scale bars = 200 µm.
(Optional pause point) Salivary cell cryo-banking
After passage 3, resuspend 80% confluent and dissociated cells from a 10 cm dish into 1 mL of salivary gland cell medium that contains 10% DMSO.
Transfer the medium with the cells into a cryogenic tube.
Freeze the vial at -80 °C fridge in a wrapped Styrofoam box.
Next day, move the vial to a LN tank for long-term storage. The cells can be safely preserved at this stage until the hPSC-derived cells are ready.
NCC induction
Prepare Geltrex-coated plates.
Thaw Geltrex at 4 °C overnight.
Dilute Geltrex with ice-cold DMEM/F12 at 1:100 dilution.
Coat the plates with Geltrex (1 mL/24-well, 2 mL/6-well plates) and seal with parafilm.
Incubate the plates at 4 °C at least overnight before use.
Plating hPSCs on day 0
Resuspend dissociated hPSCs (15 min by EDTA in order to gain single cells) in 1 mL d0, 1 media. hPSC is maintained in E8 medium. For details of hPSC replating, please also check our previous protocols [10,18].
Aspirate the Geltrex solution from the plates coated overnight.
Plate hPSCs at 125k cells/cm2 density (0.5 mL/24-well, 2 mL/6-well plates).
Incubate cells at 37 °C.
Next day (day 1), fully change the medium with NCC induction, d0, 1 media (1 mL/24-well, 3 mL/6-well plates).
On day 2, fully change the medium with NCC induction, D2–10 medium (1 mL/24-well, 3 mL/6-well plates).
From now on, feed the cells by full medium change every two days (days 2, 4, 6, 8) until day 10 (1 mL/24-well, 3 mL/6-well plates).
On day 10, differentiated NCCs should actively aggregate and form the condensed “dark ridge” structure (Figure 9) [10].
Caution: If you fail to see such structures, it is not recommended to proceed to the next step.
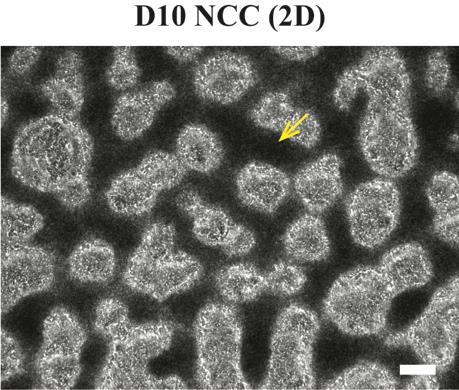
Figure 9. Cell morphology during Schwann cell precursor (SCP) differentiation. Brightfield images showing the differentiated D10 neural crest cell (NCC) “dark ridges” (indicated by the yellow arrow). Scale bar = 200 µm.
(Optional pause point) Dissociate NCCs for cryobanking
On day 10, aspirate the culture medium.
Wash cells twice with PBS (1 mL/24-well, 3 mL/6-well, and 4 mL/10 cm plates).
Add Accutase (1 mL/24-well, and 2 mL/10 cm plates).
Incubate for 20–30 min in a 37 °C incubator.
Pipette gently with a P1000 pipette 3–5 times and fully dissociate the cells.
Test if cells are ready for dissociation by gently pipetting 1–2 times. If more times are needed, incubate 5 more minutes each time and test for up to 30 min.
Transfer the Accutase and cell solution to a conical tissue culture tube that contains at least 5 times higher volume of PBS.
Centrifuge at 200× g for 5 min.
Resuspend the cells with ice-cold stem cell banker.
Count the cells.
Transfer the cells (8 × 106 cells/1 mL/vial) in ice-cold stem cell banker to cryogenic tubes.
Freeze the vial at -80 °C freezer in a wrapped Styrofoam box.
Next day, move the vial to a LN tank for long-term storage. The differentiation can be safely paused at this stage.
Dissociate NCCs for SCP differentiation
On day 10, aspirate the culture medium.
Wash cells twice with PBS (1 mL/24-well, 3 mL/6-well, and 4 mL/10 cm plates).
Add Accutase (1 mL/24-well, and 2 mL/10 cm plates).
Incubate for 20–30 min in a 37 °C incubator.
Pipette gently with a P1000 pipette 3–5 times and fully dissociate the cells.
Test if cells are ready for dissociation by gently pipetting 1–2 times. If more times are needed, incubate 5 more minutes each time and test for up to 30 min.
Transfer the Accutase and cell solution to a conical tissue culture tube that contains at least 5 times higher volume of PBS.
Centrifuge at 200× g for 5 min.
Resuspend the cells with day 10–16 medium.
Count the cells.
Replate the cells on 24-well ultra-low attachment tissue culture plates at 1 × 106/well density with SCP differentiation, D10–16 medium (0.5 mL/well).
Add day 10–16 medium on the next day (0.5 mL/well), which makes the total volume 1 mL.
Feed cells on days 13 and 15 by full medium change.
In order to keep all the cells, tilt the plate and aspirate the medium slowly. Always keep the tip on the surface of the culture medium away from the cells (Figure 10A).
On day 16, SCPs should be differentiated in suspending spheroids (Figure 10B).
Caution: If you fail to see such structures, it is not recommended to proceed to the next step.
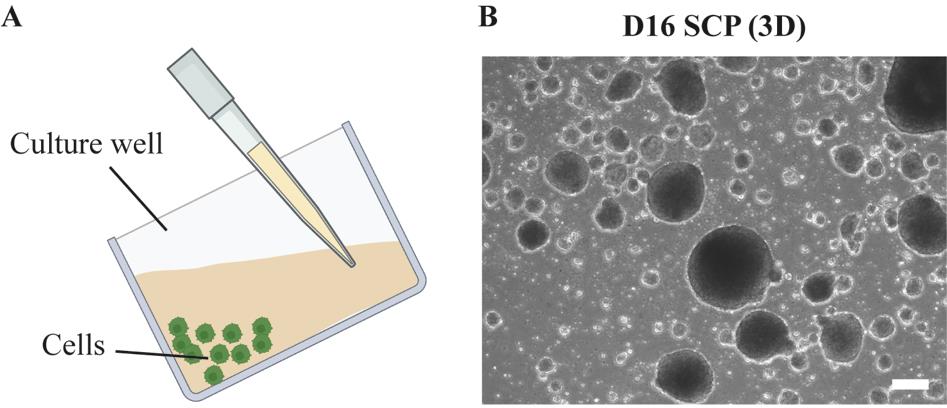
Figure 10. Cell morphology during Schwann cell precursor (SCP) differentiation. (A) Cartoon illustration of the method to change medium in suspension cultures. (B) Brightfield images showing the differentiated D16 SCP spheroids. Scale bar = 200 µm.
SCP replating for parasymN differentiation/coculture
Please make sure you have 80% confluent salivary cells at passages 3–8 before proceeding.
On day 16, tilt the plate and collect all the SCP spheroids in the culture medium into a conical tissue culture tube and fill up the tube with PBS (at least the same volume of cell suspension).
Centrifuge at 200× g for 3 min.
Aspirate the supernatant and add Accutase to the spheroids (1 mL for less than 12 wells, 2 mL for less than 24 wells).
Transfer the Accutase that contains the spheroids back to a low attachment well.
Incubate for 20–30 min in 37 °C incubator.
Gently pipette the spheroids with a P1000 pipette, transfer the cells to a conical tissue culture tube, and fill up the tube with PBS.
Centrifuge at 200× g for 5 min.
Resuspend the cells with 1 mL day 16 parasymN medium.
Count the cells.
Replate SCP cells at a density of 1 × 105/cm2 on top of 80% confluent salivary cells (passage 3–8) in a 24-well plate containing 0.5 mL parasymN differentiation medium.
Fully change the medium on the next day and every 2–3 days after (1 mL/24-well plates).
After 7–10 days, aggregated cell bodies of parasymNs should be observed on the salivary cell layer with well-developed neurites (Figure 11).
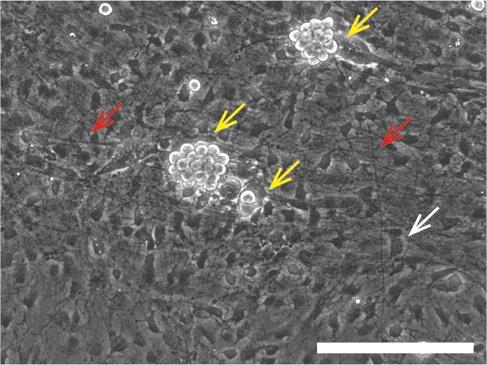
Figure 11. Cell morphology of the coculture. Brightfield images showing the cell bodies (indicated by the yellow arrows) and the axons (indicated by the red arrows) of hPSC-parasymNs on the salivary cells (indicated by the white arrow) 10 days after the coculture. Scale bar = 200 µm.
Data analysis
After 7–10 days, well-developed parasymNs should be observed on the salivary cell layer. Cell bodies of parasymNs should be clearly distinguishable from the salivary tissue, given that parasymNs will aggregate with bright and round cell bodies (Figure 11, yellow arrows). The development of neurons and their connectivity with salivary cells can be clearly observed by immunostaining with specific markers as desired (for instance, PRPH in Figure 12A) [19]. Secretory granules in salivary cells are autofluorescent and should be observable at high resolution (Figure 12B). Both total salivary cell number and granule+ salivary cells are increased with parasymNs, suggesting improved development and maturation (Figure 12C). We can also monitor Ca2+ dynamics in the salivary cells (Figure 12D). The timeline of the entire process is summarized in Figure 13.
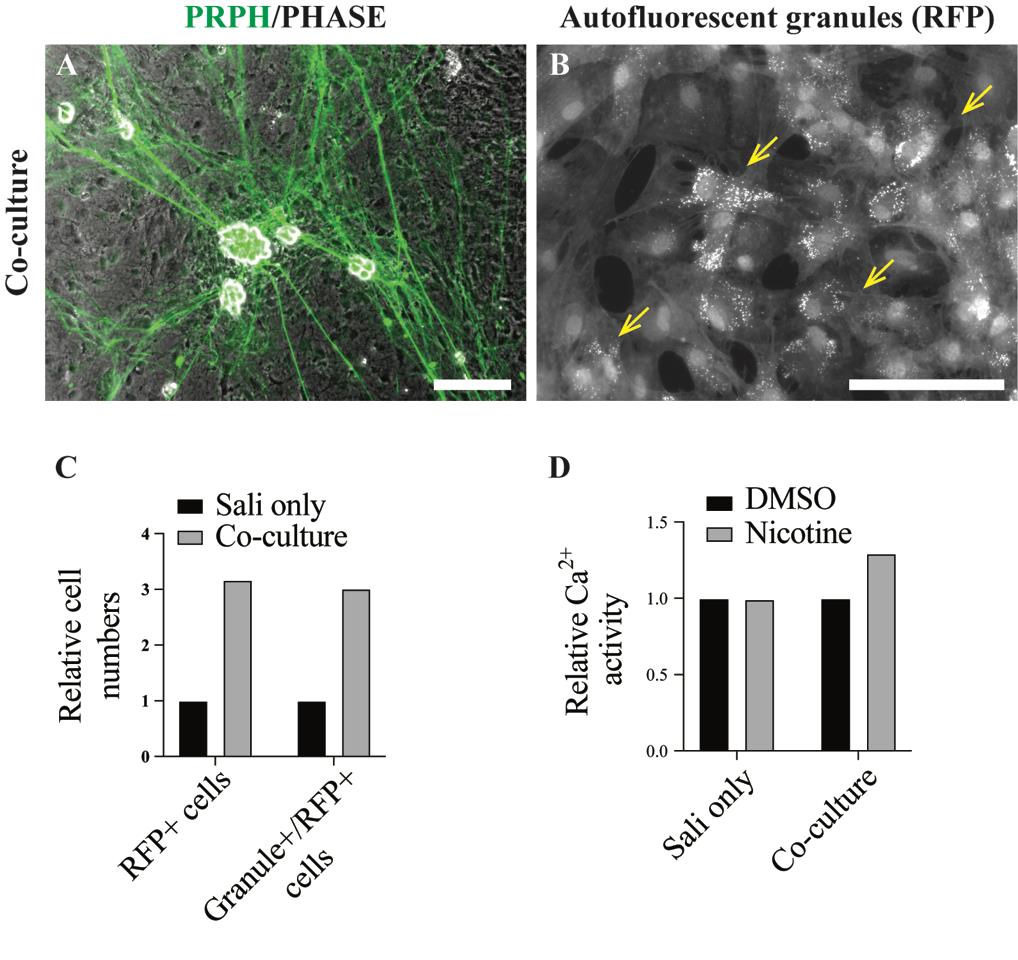
Figure 12. Sample result of functional activity of the coculture. A. Immunofluorescent staining for peripheral neural marker PRPH shows parasymNs growing on the salivary tissue with extensive neurites and aggregating cell bodies. This feature can be mostly seen in peripheral neuron cultures since it might resemble the ganglion formation in the body. B. RFP signals representing SOX10+ salivary cells as well as the autofluorescent granules. C. Representative quantification result showing the improved proliferation and maturation of the salivary tissues when cocultured with parasymNs. D. Representative Ca2+ image (using fluo-4) for the salivary cells shows the functional connectivity between parasymNs and salivary cells upon parasymN activation by nicotine in the coculture. Scale bars = 200 µm.
The replating density of D16 SCP is critical for parasymN differentiation especially in neural culture alone. However, given that current understanding suggests that there might be mutual effects on the development and maturation of cocultured cells [8], the cell density in cocultures might be adjusted; however, we have not tested this. Nevertheless, if the density is too high, the chance that the cells detach after coculture will be higher. It is also worth mentioning that, while hPSC-derived parasymNs allow an unlimited neuron source for research, the nature of small amounts and limited passage numbers of primary salivary tissues still restrain the scale-up option of this coculture model system. Establishing a coculture model with both hPSC-derived parasymNs and hPSC-derived salivary tissues would be an important future task.
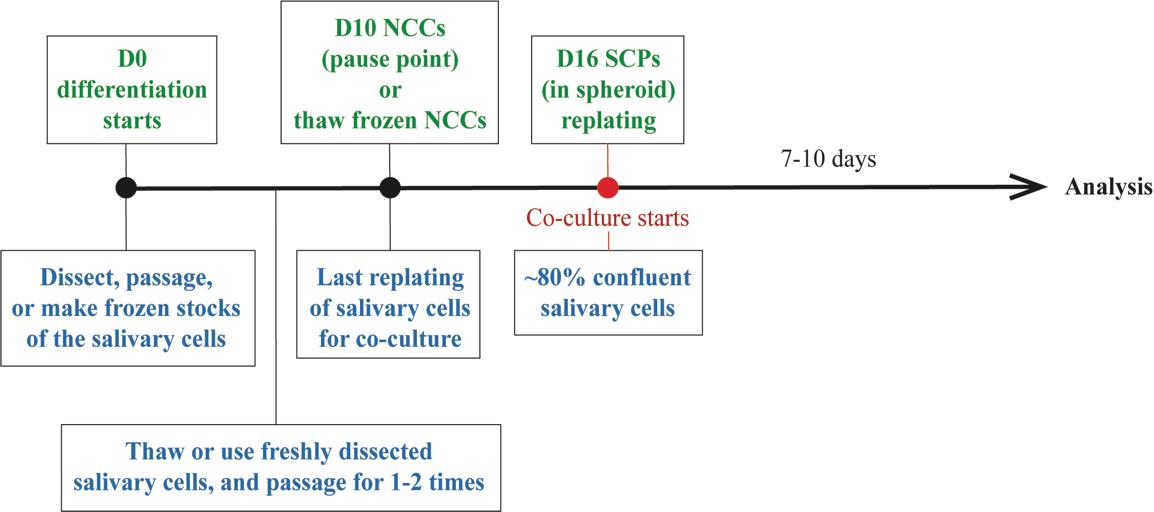
Figure 13. Timeline of the coculture
Validation of protocol
This protocol or parts of it has been used and validated in the following research article(s):
Wu et al. [12]. Parasympathetic neurons derived from human pluripotent stem cells model human diseases and development. Cell Stem Cell (Figure 6, panel H, and supplementary figure 9).
Yu et al. [16]. The duct of von Ebner’s glands is a source of Sox10+ taste bud progenitors and susceptible to pathogen infections. Frontiers in Cell and Developmental Biology (Figure 1, panel A and B).
General notes and troubleshooting
General notes
Efficient and successful early NCC induction for the first 10 days of differentiation is critical. For more detailed methods and tips, please also see our previous protocols [10,18].
It is always recommended to replate a neuron-only group and a salivary gland-only group in parallel to the cocultures to validate the neuron differentiation efficiency and functionality as well as the salivary gland cell integrity of each batch. When differentiating parasymNs alone, coat the plate with PO (15 μg/mL in DPBS)/LM (2 μg/mL in DPBS)/FN (2 μg/mL in DPBS).
Troubleshooting
Problem 1: D16 SCP spheroid is poorly formed.
Possible cause: D10 NCC is not properly differentiated (very few or no NC ridges can be observed).
Solution: It is critical to make sure that each stage of the differentiation, including D0 hPSC and D10 NCC, has passed the quality control steps as previously described [10]. If NCC differentiation is not ideal, a BMP4 titration test is highly recommended, as detailed in [18]. In general, concentrations between 0.2 and 1 ng/mL are ideal. In our case, for example, we had to drop the concentration from 1 to 0.4 ng/mL due to the batch-to-batch variability of BMP4.
Problem 2: ParasymNs in the cocultures do not regulate the activity of the salivary tissues.
Possible cause: The parasymN or salivary cell batch is bad.
Solution: Only use salivary cells between passages 3 and 8. Always replate a neuron-only group in parallel to the coculture to validate the neuron differentiation efficiency and functionality of each batch.
Problem 3: Difficulty in injecting tongue tissue separation solution into subepithelial space.
Possible cause: Incorrect injection technique or improper handling of the tongue.
Solution: Practice the injection technique to ensure accurate delivery of solution. Use a dissecting microscope for better visualization and precision. Be careful not to poke and injure the tongue epithelium; otherwise, the solution will leak, leading to incomplete enzyme digestion.
Problem 4: Incomplete or over-enzyme digestion for tongue tissue separation.
Possible cause: Insufficient enzyme incubation time, incorrect enzyme concentration, or improper injection of enzyme. Solution: Verify the concentration and activity of Collagenase A and Dispase II. Ensure proper mixing and incubation at the correct temperature for the specified duration.
Problem 5: Primary cell clumping or low viability.
Possible cause: Over-digestion of tissues for the separation or excessive mechanical stress during pipetting.
Solution: Control the incubation time ≤ 30 min at 37 °C after the injection of tongue tissue separation solution. Agitate gently during the incubation with salivary gland cell dissociation solution. Consider optimizing the trypsin-EDTA concentration and incubation time.
Acknowledgments
This work was supported by 1R01NS114567-01A1 to NZ and R21DC018910 and R21DC018089 to HXL. This protocol was adapted from Wu et al. [12] and Yu et al. [15].
Competing interests
The authors declare no competing interests.
Ethical considerations
This study employed human embryonic stem cell lines (WA09), the use of which by the Zeltner lab was approved by WiCell. All iPSCs employed in this work were reprogrammed from human samples obtained through the public repository Coriell Research Institute. The use of animals was approved by the Institutional Animal Care and Use Committee at The University of Georgia (AUP# A2022 06-030-A6) and complied with the National Institutes of Health Guidelines for the care and use of animals for research.
References
- McCorry, L. K. (2007). Physiology of the autonomic nervous system. Am J Pharm Educ. 71(4): 78.
- Lefcort, F. (2020). Development of the Autonomic Nervous System: Clinical Implications. Semin Neurol. 40(5): 473–484.
- Scott-Solomon, E., Boehm, E. and Kuruvilla, R. (2021). The sympathetic nervous system in development and disease. Nat Rev Neurosci. 22(11): 685–702.
- Muo, E. C., Cardona, J. J., Chaiyamoon, A., Iwanaga, J. and Tubbs, R. S. (2023). The Parasympathetic Root of the Submandibular Ganglion: A Review. Cureus. 15(1): e33775.
- Takano, T., Wahl, A. M., Huang, K. T., Narita, T., Rugis, J., Sneyd, J. and Yule, D. I. (2021). Highly localized intracellular Ca2+ signals promote optimal salivary gland fluid secretion. eLife. 10: e66170.
- Frith, T. J. R. and Tsakiridis, A. (2019). Efficient Generation of Trunk Neural Crest and Sympathetic Neurons from Human Pluripotent Stem Cells Via a Neuromesodermal Axial Progenitor Intermediate. Curr Protoc Stem Cell Biol. 49(1): e81.
- Kirino, K., Nakahata, T., Taguchi, T. and Saito, M. K. (2018). Efficient derivation of sympathetic neurons from human pluripotent stem cells with a defined condition. Sci Rep. 8(1): 12865.
- Oh, Y., Cho, G. S., Li, Z., Hong, I., Zhu, R., Kim, M. J., Kim, Y. J., Tampakakis, E., Tung, L., Huganir, R., et al. (2016). Functional Coupling with Cardiac Muscle Promotes Maturation of hPSC-Derived Sympathetic Neurons. Cell Stem Cell. 19(1): 95–106.
- Wu, H. F. and Zeltner, N. (2019). Overview of Methods to Differentiate Sympathetic Neurons from Human Pluripotent Stem Cells. Curr Protoc Stem Cell Biol. 50(1): e92.
- Wu, H. F. and Zeltner, N. (2020). Efficient Differentiation of Postganglionic Sympathetic Neurons using Human Pluripotent Stem Cells under Feeder-free and Chemically Defined Culture Conditions. J Vis Exp. (159). doi.org/10.3791/60843.
- Saito-Diaz, K., Wu, H. F. and Zeltner, N. (2019). Autonomic Neurons with Sympathetic Character Derived From Human Pluripotent Stem Cells. Curr Protoc Stem Cell Biol. 49(1): e78.
- Wu, H. F., Saito-Diaz, K., Huang, C. W., McAlpine, J. L., Seo, D. E., Magruder, D. S., Ishan, M., Bergeron, H. C., Delaney, W. H., Santori, F. R., et al. (2024). Parasympathetic neurons derived from human pluripotent stem cells model human diseases and development. Cell Stem Cell. 31(5):734–753.e8.
- Espinosa-Medina, I., Saha, O., Boismoreau, F., Chettouh, Z., Rossi, F., Richardson, W. D. and Brunet, J. F. (2016). The sacral autonomic outflow is sympathetic. Science. 354(6314): 893–897.
- Proctor, G. B. and Carpenter, G. H. (2014). Salivary secretion: mechanism and neural regulation. Monogr Oral Sci. 24: 14–29.
- Yu, W., Ishan, M., Yao, Y., Stice, S. L. and Liu, H. X. (2020). SOX10-Cre-Labeled Cells Under the Tongue Epithelium Serve as Progenitors for Taste Bud Cells That Are Mainly Type III and Keratin 8-Low. Stem Cells Dev. 29(10): 638–647.
- Yu, W., Kastriti, M. E., Ishan, M., Choudhary, S. K., Rashid, M. M., Kramer, N., Do, H. G. T., Wang, Z., Xu, T., Schwabe, R. F., et al. (2024). The duct of von Ebner’s glands is a source of Sox10+ taste bud progenitors and susceptible to pathogen infections. Front Cell Dev Biol. 12: e1460669.
- Venkatesan, N., Boggs, K. and Liu, H. X. (2016). Taste Bud Labeling in Whole Tongue Epithelial Sheet in Adult Mice. Tissue Eng Part C Methods. 22(4): 332–337.
- Wu, H. F., Art, J., Saini, T. and Zeltner, N. (2024). Protocol for generating postganglionic sympathetic neurons using human pluripotent stem cells for electrophysiological and functional assessments. STAR Protoc. 5(2): 102970.
- Wu, H., Saito-Diaz, K., Huang, C. W., McAlpine, J. L., Seo, D. E., Magruder, D. S., Ishan, M., Bergeron, H. C., Delaney, W. H., Santori, F. R., et al. (2024). Parasympathetic neurons derived from human pluripotent stem cells model human diseases and development. Cell Stem Cell. 31(5): 734–753 e738.
Article Information
Publication history
Received: Jul 19, 2024
Accepted: Sep 9, 2024
Available online: Oct 13, 2024
Published: Nov 5, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Wu, H. F., Ishan, M., Rashid, M. M., Liu, H. X. and Zeltner, N. (2024). Novel Cross-Species Salivary Gland-Parasympathetic Neuron Coculture System. Bio-protocol 14(21): e5101. DOI: 10.21769/BioProtoc.5101.
Category
Stem Cell > Pluripotent stem cell > Cell differentiation
Cell Biology > Cell engineering > Tissue engineering
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










