- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Extraction of Bacterial Membrane Vesicle and Phage Complex by Density Gradient Ultracentrifugation
(*contributed equally to this work) Published: Vol 14, Iss 16, Aug 20, 2024 DOI: 10.21769/BioProtoc.5050 Views: 835
Reviewed by: Alba BlesaNuttavut KosemManousos E. Kambouris

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
General Maintenance and Reactivation of iSLK Cell Lines
Ariana C. Calderón-Zavala [...] Ekaterina E. Heldwein
Jun 5, 2025 710 Views
Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 871 Views
Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 838 Views
Abstract
The bacterial membrane vesicles (MVs) are non-replicative, nanoscale structures that carry specific cargos and play multiple roles in microbe–host interactions. An appropriate MV isolation method that mimics complex pathogen infections in vivo is needed. After bacterial MVs extraction, flagella or pili can be frequently observed along with MVs by transmission electron microscope (TEM). Recently, MVs from Pseudomonas aeruginosa were found to coexist with Pf4 phages, and this MV–phages complex exhibited a different impact on host cell innate immunity compared with MVs or phages solely. The presence of this MVs–phages complex simulates the real condition of complex pathogen infections within the host. This protocol outlines the extraction of the MVs and Pf4 phages complex of P. aeruginosa PAO1, including the respective isolation and qualification approaches. Our step-by-step bacterial MVs–phages complex extraction protocol provides valuable insights for further studying microbe–host cell interactions and the development of novel phage therapies.
Key features
• Detailed density gradient extraction procedures of MVs–phages complex
• TEM, plaque assay, and PCR to verify the coexistence of MVs and phages
• The obtained MVs–phages complex can be used for exploring phage–microbe–host cell interactions
Keywords: Pseudomonas aeruginosaGraphical overview
Background
Pseudomonas aeruginosa is an opportunistic pathogen that often causes serious hospital-acquired infections and can lead to a variety of infectious diseases including pneumonia, wound infections, and septicemia [1]. Due to the high mortality rate of these infections, P. aeruginosa was recognized as one of the most life-threatening bacteria and listed as a priority pathogen for research and development of new antibiotics by the World Health Organization [2].
The Pf phage, a filamentous inovirus specifically associated with P. aeruginosa, is a type of temperate phage, which can be classified from Pf1 to Pf8. Approximately half of P. aeruginosa isolates harbor Pf phage operons within their genomes [3,4]. Pf phages are about 6–7 nm in diameter and vary in length from 0.8 to 2 µm, with a single-stranded DNA (ssDNA) as the genetic material packaged within a helical filamentous structure major coat protein CoaB [3]. Of these, Pf4 phage is an important member, and its genome is integrated into the P. aeruginosa PAO1 as the prophage. Thus, Pf4 phage can emerge inside the bacteria [5]. The core genes of the Pf4 phages mainly include xisF4, PA0720, PA0723, PA0724, and PA0726, which encode excisionase, single-stranded DNA binding protein, major coat protein CoaB, minor coat protein CoaA, and morphogenesis protein, respectively [3,5]. A recent study showed that Pf4 phages can be transmitted between bacteria and have various effects on P. aeruginosa, including reducing twitching motility, altering biofilm production, and affecting the release of virulence factors, which are important in the pathogenesis of P. aeruginosa [6–8]. However, how Pf4 is transmitted has remained elusive.
Bacterial membrane vesicles (MVs) are structures produced by bacteria with a diameter ranging from 40 to 400 nm that carry specific cargo and travel a long distance (centimeter scale) to play essential roles in cell–cell communications [9,10]. Gram-negative bacteria MVs could be divided into two types: B (blebbing)-type vesicles, which are formed via blebbing from the outer membrane, and E (explosive)-type vesicles, which are formed due to explosive cell lysis [11,12]. Studies have suggested that P. aeruginosa E-MVs can be formed under certain stresses and bacteriophage lysis conditions induced by Pf4 phages [3].
Many previous works observed that after the crude MV extraction step, flagella can also be observed by TEM [13]. A recent study investigated the combined effects of P. aeruginosa MVs with phages upon the recognition function of the host’s innate immune system. In a mouse model of acute lung infection, treatment with MVs-Pf4 phages complex resulted in reduced infiltration of neutrophils in the lung alveoli, thus suppressing the occurrence of inflammatory reactions [3].
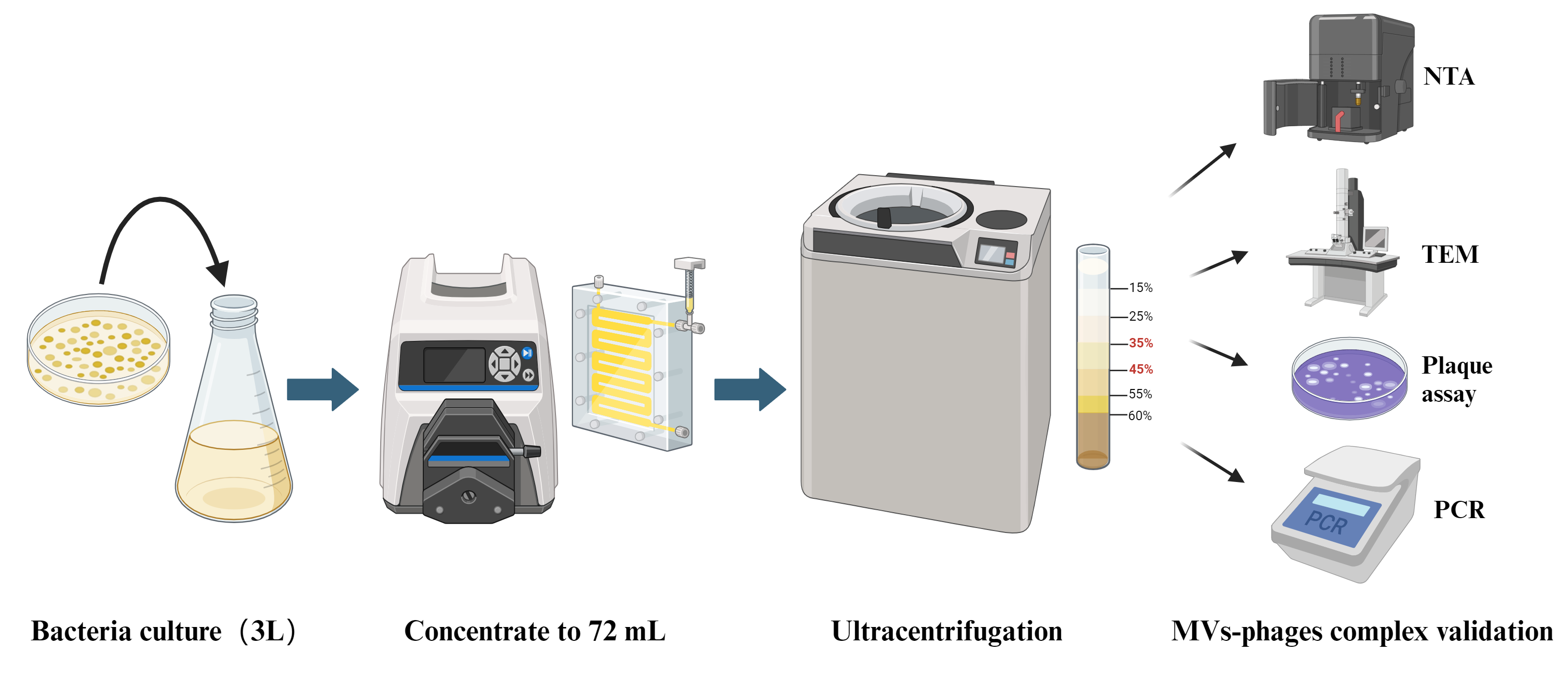
In this protocol, we establish a bacterial MVs–phages co-extraction platform from P. aeruginosa PAO1 as a model organism. The MVs–phages extract effectively mimics the complex MVs–phage coexistence condition in vivo within the host during pathogen infections. Overall, this is an optimized method for extracting the MV–phage complex based on density gradient centrifugation (Figure 1), with feasibility verified through TEM, PCR, and plaque assays. It will provide a reliable protocol for studying bacteriophage–microbe–host cell interactions.
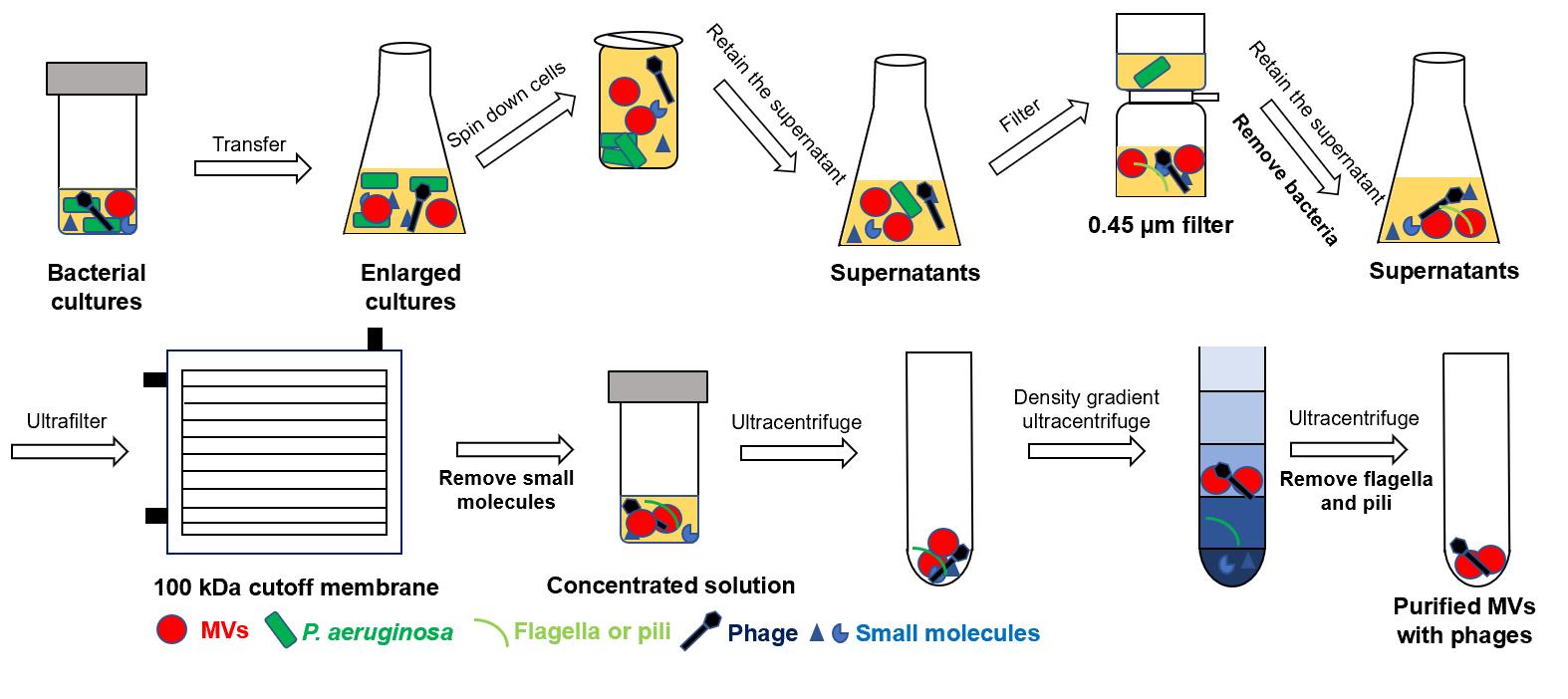
Figure 1. Flowchart of MV–phage complex extraction. The bacterial culture is centrifuged to retain the supernatant. After filtering and ultrafiltering, bacteria and small molecules can be removed. After density gradient ultracentrifuging, flagella and pili can be removed. Then, purified MVs–phage samples can be obtained.
Materials and reagents
The following materials and equipment are recommended for this protocol, but alternatives from other sources can also be used when proven to be equivalent.
Biological materials
P. aeruginosa PAO1 strain (strains are preserved at -80 °C in our laboratory)
P. aeruginosa PAO1 ΔPf4 strain (strains are preserved at -80 °C in our laboratory)
Opti-Prep gradient medium (Sigma-Aldrich, catalog number: D1556)
Phosphate-buffered saline (PBS) (Servicebio, catalog number: G4202)
2% Phosphotungstic acid solution (Acmec, catalog number: AG1599)
LB liquid medium (Servicebio, catalog number: G3103)
Premix TaqTM Version 2.0 (Takara, catalog number: R004Q)
1% agarose gel (Baygene, catalog number: 192255)
DNA marker (Coolaber, catalog number: D2000)
Pancreatic digest of casein (peptone) (Beyotime, catalog number: ST800)
Soybean digest (Sigma-Aldrich, catalog number: C7210)
NaCl (Dogesce, catalog number: Sodium Chloride)
Agar (Dogesce, catalog number: Agar)
CaCl2 (Macklin, catalog number: C915443-500g)
MgCl2 (Macklin, catalog number: M813766-500g)
Mitomycin C (AbMole, catalog number: M5791)
GelRed 10,000× gel staining solution (I-PRESCI, catalog number: P202-01)
TSB culture medium (see Recipes)
1.5% LB agar solid medium (see Recipes)
0.75% LB agar semi-solid medium (see Recipes)
TSB culture medium
The composition of the medium includes distilled water, 1.5% pancreatic digest of casein (peptone), 0.5% soybean digest, and 0.5% NaCl.
1.5% LB agar solid medium
100 mL LB liquid medium with 1.5 g of agar.
0.75% LB agar semi-solid medium
100 mL LB liquid medium with 0.75 g agar, 1 mM CaCl2, and 1 mM MgCl2.
Laboratory supplies
LB agar plate (Servicebio, catalog number: G3104-0910)
50 mL cell culture tube (Bioland, catalog number: ATS05-12-50)
13.2 mL ultracentrifuge tube (Beckman, catalog number: 344059)
1.5 mL centrifuge tube (Eppendorf, catalog number: 0030108051)
PCR tube (Eppendorf, catalog number: 0030124359)
500 mL filter flask with 0.45 μm filter (Biosharp, catalog number: BS-500-XT)
Glow-discharged 200-mesh carbon grid (Beijing Zhongjingkeyi Technology, catalog number: BZ11022B)
250 mL centrifuge bottle (Beckman, catalog number: 356011)
Equipment
Intelligent high-efficiency centrifuge (Avanti, model: JXN-26)
Masterflex easy-load machine (Sartorius AG, model: VFA013)
100 KD Vivaflow membranes (Sartorius AG, model: VF20H4)
Ultracentrifuge machine (Beckman, model: Optima XE-100)
High-pressure sterilization pot (Yamato, model: SQ810C)
Constant temperature shaker (Shanghaiminquanyiqi, model: MQT-60R)
Tecan Spark 10 M plate reader (Tecan Group Ltd., model: Tecan Spark 10 M)
Transmission electron microscope (Hitachi, model: HT7700)
Nano Sight nanoparticle tracking analysis machine (Malvern Instruments Ltd, model: NS300)
Procedure
Preparation of the culture medium
Prepare 3 L of TSB culture medium (see Recipes).
Note: Adjust volume depending on bacteria. We use 3 L for one sample to obtain abundant MVs. The expected protein quantification concentration of the post-enriched MVs solution is 200 ng/μL.
Sterilize the medium in a high-pressure autoclave (121 °C, 21 min).
Bacterial culture
Select P. aeruginosa PAO1 from the -80 °C refrigerator, streak on a 1.5% solid LB plate, and culture upside down overnight at 37 °C.
Using a 50 mL cell culture tube, inoculate a single bacterial colony into the sterilized 20 mL of TSB culture medium and incubate it on a shaking bed at 37 °C until the OD600nm reaches 0.6.
Sub-culture bacteria into 3 L of TSB culture medium and culture on a shaking bed at 37 °C overnight until it reaches the exponential phase (OD600nm > 1.0).
Extraction of MV and filamentous structures complex
Transfer all the above-mentioned cultures to 250 mL centrifuge tubes and centrifuge at 10,000× g for 15 min at 4 °C.
Filter the supernatant through the 500 mL filter flask with the 0.45 μm filter to remove any remaining bacterial cells.
Transfer the above solution to a 3 L conical flask. Connect 100 KD Vivaflow membranes to the outlet end of the Masterflex Easy-Load system, start the system, and concentrate the solution to 72 mL.
Note: This is a rate-limiting step: 2 L of solution normally needs 1 h to be concentrated.
Transfer the concentrate into ultracentrifuge tubes with 12 mL in each tube. Ultracentrifuge concentrated supernatant at 50,000× g for 2 h at 4 °C to precipitate crude MVs. Remove the supernatant.
Add the Opti-Prep gradient medium into the ultracentrifuge tube and then add the crude extracted MV to the top of the solution. Perform density gradient centrifugation purification of the crude MVs using Opti-Prep gradient medium with a gradient range of 15%–60% at 4 °C and centrifuge at 100,000× g for over 16 h overnight. The purified MVs will be located in the 35%–45% density layer (Figure 2).
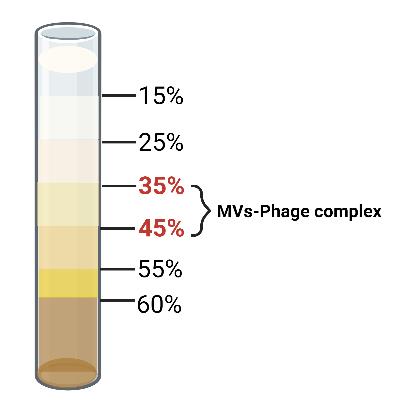
Figure 2. Stratification of the MVs-phages complex in the Opti-prep medium. After centrifugation at 4 °C for 16 h at 100,000× g, the MVs–phages complex is distributed in the 35%–45% layer of the Opti-prep gradient. Purified MVs can be taken using a 1 mL syringe.Take the purified MVs using a 1 mL syringe into a 1.5 mL centrifuge tube and wash them continuously by ultracentrifugation with 4 °C 1× PBS at 50,000× g for 2 h at 4 °C.
Notes: The 1.5 mL centrifuge tubes must be soaked in 75% alcohol overnight.
Resuspend the purified MVs in 1 mL of PBS and store at 4 °C.
Validation of the MV–phage coexistence
Nanoparticle tracking analysis (NTA) to assess the quality of MVs
The quantification and size characterization of the MVs were analyzed using the Nano Sight NS300 for NTA.
Note: Nano Sight NS300 is an instrument used for size analysis and counting of nanoparticles and biological particles. It uses the laser to track the movement of particles in a fluid and analyzes these movement trajectories to calculate the size, concentration, and distribution of the particles.
MV samples were diluted (500×) in PBS to obtain a concentration within the recommended measurement range (1–10 × 108 particles/mL). The size and concentration measurements were conducted using the 448 nm laser in scatter mode.
Using a 1 mL syringe, the sample was injected into the instrument and videos were captured in triplicate for 30 s. The mean values for size and concentration were analyzed using NanoSight (NTA software, version 3.0).
Coexistence of MV and phage visualized by negative staining TEM
Incubate 5 μL of the purified MVs on a glow-discharged 200-mesh carbon grid for 1 min to allow binding.
Contrast the grids with a 2% phosphotungstic acid solution.
Fit the microscope with a high-sensitivity real-time charge-coupled device camera for image capture.
View the grids on an HT7700 transmission electron microscope operating at 100 kV.
Plaque assay to verify the coexistence of phages in bacterial MVs
Pour 25 mL of sterilized 1.5% LB medium with agar into a Petri dish and let it solidify as the underlayer. Take another sterilized 25 mL of LB semi-solid medium (containing 0.75% agar), add 600 μL of freshly cultured ΔPf4 bacteria (OD600nm = 0.6) into this medium, and mix thoroughly. Then pour onto the solidified underlayer as the upper layer.
Prepare the bacteriophage: add 0.5 μg/mL mitomycin C to the culture medium and culture the bacteria overnight. Take 1 mL of the culture fluid for centrifugation and filter the supernatant twice through a 0.22 μm filter.
The bacteriophages to be tested should be diluted in sterile PBS in a 10-fold serial dilution. After each dilution, take 5 μL of the bacteriophage solution, spot it onto the surface of a bacterial semi-solid agar plate, and allow it to dry.
Incubate the plate inverted at 37 °C overnight. The following day, count and analyze the plaques formed.
PCR validation of the coexistence of Pf4 phages in bacterial MVs
Design primers of Pf4 phages for the target genes PA0720, PA0724, and PA0726 based on the gene sequence of the Pf4 bacteriophage using SnapGene software (Table 1). Synthesize the primers and dissolve them.
Note: When validating the binding of other types of bacteriophages in experiments, it is necessary to design primers for specific genomic loci of the bacteriophage according to the actual situation.
Table 1. Primer sequences for PA0720, PA0724, and PA0726
PA0720 PA0724 PA0726 Forward primer ACGAACACCCGTGGTTGGCT AAGGCGTAACGGGTGCTCTG CTCGATCAGATCATCGCCTT Reverse primer TAGGCCTGTTGCCATGCGAG GCGGCATACATGCTGCGGAT TGGTGTCGTCGAAGAACAAC Using 1 μL of purified MVs as templates, perform PCR by adding primers. The PCR reaction system is as follows (Table 2 and Table 3).
Table 2. PCR reaction master mix
Reagent Amount Premix TaqTM Mix 25 μL Template 1 μL Forward primer 2 μL Reverse primer 2 μL ddH2O 20 μL Table 3. PCR cycling conditions
Steps Temperature Time Cycles Initial denaturation 98 °C 10 min 1 Denaturation 98 °C 10 s
35 cyclesAnnealing 55 °C 30 s Extension 72 °C 1 Kb/min Final extension 72 °C 5 min 1 Hold 4 °C Hold After the reaction, prepare 50 mL of 1% agarose gel, heating it in a microwave until completely dissolved. Let it cool down to around 50 °C, then add 5 μL of GelRed 10,000× gel staining solution, mix well, and pour the mixture into a gel tray.
Load the samples and DNA marker onto the gel and perform electrophoresis at a constant voltage of 164 V for 20 min using an electrophoresis apparatus.
Visualize the gel using a gel imaging system for observation.
Data analysis
In this protocol, MVs require stringent validation and quality control measures. To this end, an initial step focuses on the measurement and analysis of the size of the extracted MVs, which is an essential aspect of assessing the quality of the MV extraction process. Techniques such as TEM, plaque assay, and PCR are performed to verify the coexistence of MVs and Pf4 phage. Herein, we analyze the data obtained to provide experimental references according to Liu et al. [14].
NTA test analysis of MVs size and concentration
Using the NTA method, the size and concentration of the extracted MVs can be measured to assess the quality of the extracted P. aeruginosa MVs. Results show that the (mean) size and concentration of MVs in PAO1 WT are 67.19 nm and 2.03 × 1010 particles/mL, respectively (Figure 3).
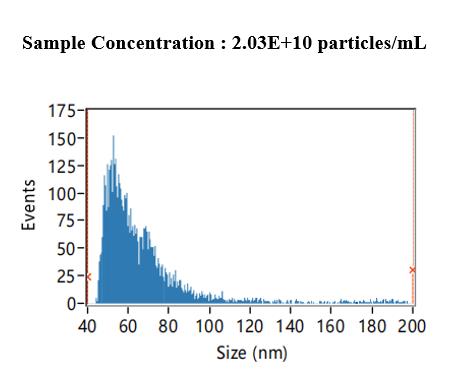
Figure 3. Bacterial membrane vesicles (MVs) concentration and size measured by the nanoparticle tracking analysis (NTA) method. Based on the NTA measurement and software analysis results, we can conclude that the concentration of the measured sample is 2.03 × 1010 particles/mL, with sizes distributed mainly between 30 and 80 nm and with a mean size of 67.19 nm.TEM observation of the coexistence of MVs and filamentous structures
We used negative staining TEM to observe the structures of the extracted complex. MVs, flagella, and phages can be observed directly through this method (Figure 4). TEM results clearly show that MVs are nearly 100–200 nm in size and comprise spherical structures harboring membrane bilayers. However, it is hard to distinguish whether the filamentous structures are phage or flagella from the outside structure. Thus, we performed further verifications to confirm whether phages were included.
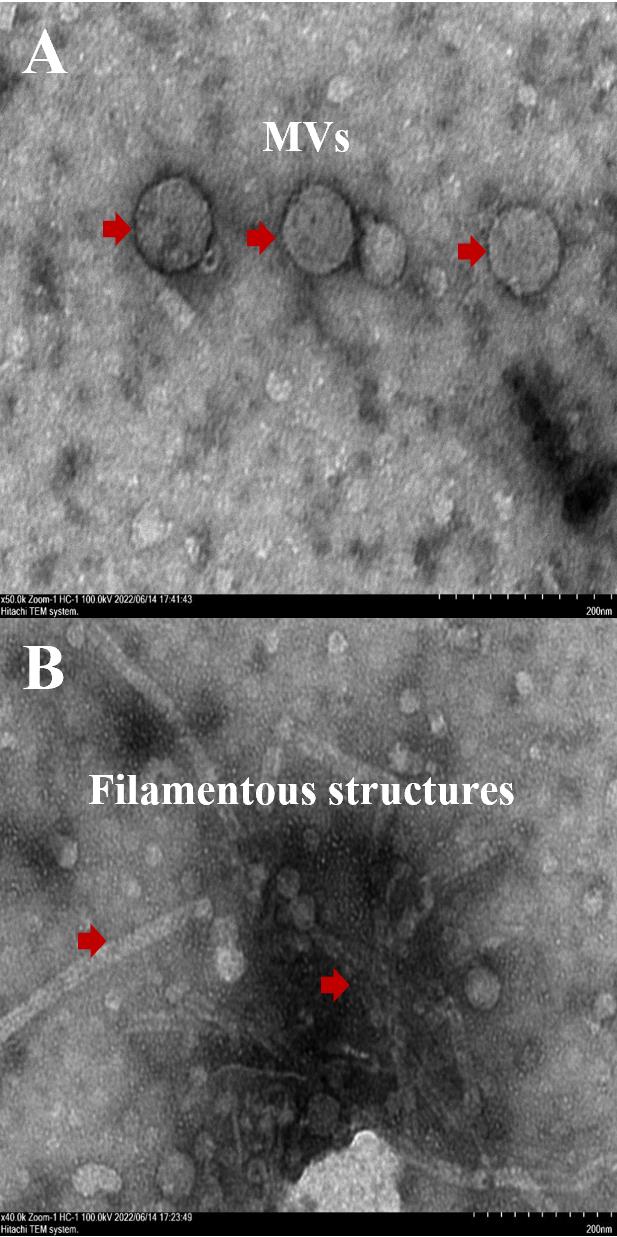
Figure 4. TEM observation to show the coexistence of membrane vesicles (MVs) and filamentous structures. A. In the image, white spherical structures with a size of approximately 100–200 nm can be observed, as indicated by the red arrow, which is the extracted MVs. B: Multiple filamentous structures can be observed in the image, as indicated by the red arrow, which is a mixture of flagella and bacteriophages.Plaque assay to verify the coexistence of phages and MVs
We utilized the supernatant of the bacterial strain culture fluid as a positive control and the supernatant of the ΔPf4 strain culture fluid as a negative control, demonstrating the presence of bacteriophages in the extracted MVs. The concentration of bacteriophages in the original sample is expressed in plaque-forming units (PFU), and the formula is PFU = (The number of plaques * Dilution factor)/ the volume of the inoculum.
Quantification results show that the number of plaque-forming units (PFU) dropped from 5.2 × 104 in the P. aeruginosa phages to 600 in PAO1 MVs, thereby showing the coexistence of phages and MVs (Figure 5).
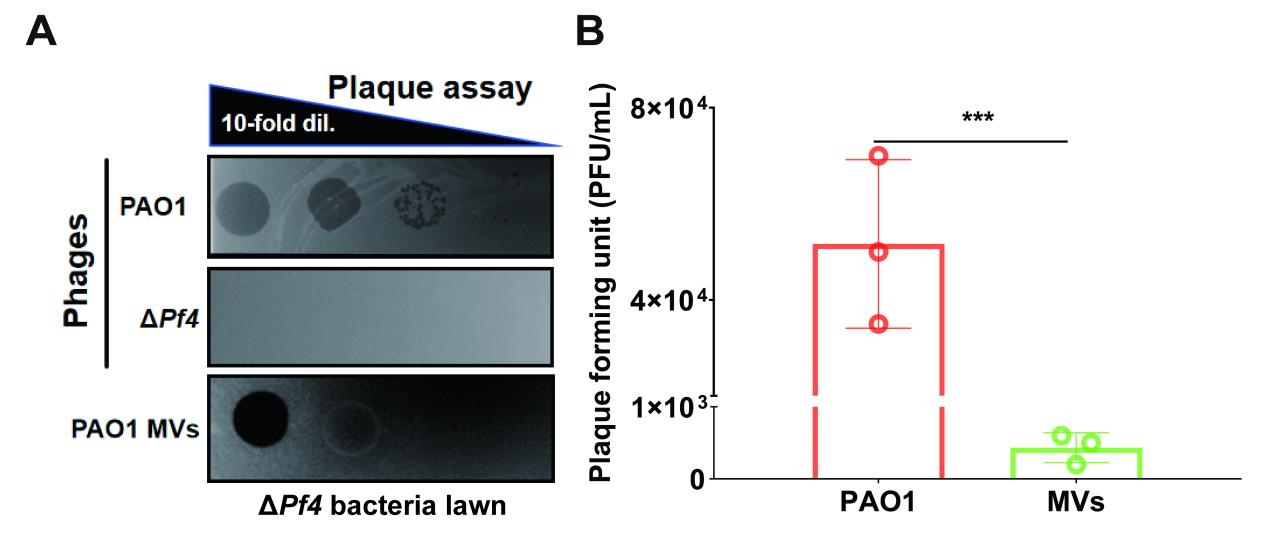
Figure 5. Plaque assay to verify the coexistence of phages in bacterial MVs. A. Plaque assay of PAO WT, ΔPf4 phages, and PAO1 MVs on the ΔPf4 bacteria lawn. B. Quantification of PFUs in PAO1 WT bacteria and PAO1 MVs. Data are presented as means ± SD (n =3, ***P ≤ 0.001, Mann-Whitney U test).PCR verification of the coexistence of Pf4 phages and MVs
In this experiment, we selected three characteristic gene fragments, PA0720, PA0724, and PA0726, as important indicators for detecting the presence of the Pf4 bacteriophage. After conducting PCR on the purified P. aeruginosa MVs, we detected the characteristic gene fragments of the Pf4 phage, PA0720, PA0724, and PA0726, which further showed that P. aeruginosa MVs seemed to coexist with Pf4 phage (Figure 6).
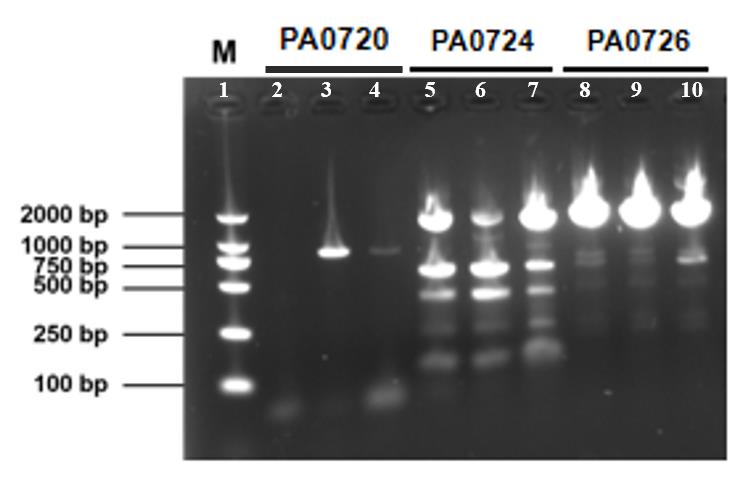
Figure 6. PCR verification of coexistence of Pf4 phages in bacterial membrane vesicles (MVs). Lane 1: DNA marker. The size of PA0720 is 816 bp, and enrichment is visible in lanes 3 and 4; the size of PA0724 is 399 bp, and enrichment is visible in the 250–500 bp region of lanes 5–7; the size of PA0726 is 1937 bp, and significant enrichment is visible around 2,000 bp in lanes 8–10. This confirms the presence of the Pf4 phage in the extracts.
Validation of protocol
The whole procedure has been validated in Pennetzdorfer et al [3].
General notes and troubleshooting
In this experiment, we chose P. aeruginosa PAO1 as the experimental subject. When using the protocol to extract MV–phage complexes from other bacteria, it is important to select a more suitable culture medium.
In the bacterial culture step, it is necessary to cultivate the bacteria to the exponential growth phase and monitor that the OD600 value exceeds 1.0 to obtain a sufficient number of bacteria and MVs. If the OD600 value is too low, it may reduce the yield of MVs. If the MV extraction concentration is low, it may be helpful to attempt further bacterial expansion.
Density gradient centrifugation is the most critical step in this experiment, requiring careful attention to centrifugation speed, time, and temperature. Insufficient centrifugation time can lead to incomplete layer separation and result in mixed impurities in the extract. When using a syringe to extract MVs–phage complexes from the 35%–45% layer, caution is necessary to avoid contamination from other layers. If mixing between layers occurs during this step, it is necessary to repeat this procedure.
For experimental rigor, we employed NTA, TEM, plaque assay, and PCR to confirm the coexistence of extracted MVs with Pf4. During practical operations, depending on needs and lab equipment, you can select necessary inspection methods to quality-check the extracted complex.
After extraction, the protein concentration of the extracted MVs can be determined by the BCA method for preliminary quality inspection. Following the complete protocol procedure, the expected protein concentration is approximately 200 ng/μL. If the measured protein concentration is too low, it indicates that the quantity of extracted MVs is insufficient, and it may be necessary to consider repeating the bacterial culture steps to achieve a higher initial bacterial concentration.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2022YFC2304700); the National Natural Science Foundation of China (32300068, 32270196, 91951204, 32200155, 32200053, and 32300060); Guangdong Basic and Applied Basic Research Foundation (2019A1515110640 and 2020A1515010316); Shenzhen Science and Technology Program (KQTD20200909113758004, 2022303002); and Guangdong Pearl River Talent Plan.
Competing interests
The authors declare no competing interests.
References
- Curran, C. S., Bolig, T. and Torabi-Parizi, P. (2018). Mechanisms and Targeted Therapies for Pseudomonas aeruginosa Lung Infection. Am J Respir Crit Care Med. 197(6): 708–727. https://doi.org/10.1164/rccm.201705-1043SO.
- Thi, M. T. T., Wibowo, D. and Rehm, B. H. A. (2020). Pseudomonas aeruginosa Biofilms. Int J Mol Sci. 21(22). https://doi.org/10.3390/ijms21228671.
- Pennetzdorfer, N., Popescu, M. C., Haddock, N. L., Dupuy, F., Kaber, G., Hargil, A., Johansson, P. K., Enejder, A. and Bollyky, P. L. (2023). Bacterial outer membrane vesicles bound to bacteriophages modulate neutrophil responses to bacterial infection. Front Cell Infect Microbiol. 13: 1250339. https://doi.org/10.3389/fcimb.2023.1250339.
- Salmond, G. P. and Fineran, P. C. (2015). A century of the phage: past, present and future. Nat Rev Microbiol. 13(12): 777–786. https://doi.org/10.1038/nrmicro3564.
- Secor, P. R., Burgener, E. B., Kinnersley, M., Jennings, L. K., Roman-Cruz, V., Popescu, M., Van Belleghem, J. D., Haddock, N., Copeland, C., Michaels, L. A., et al. (2020). Pf Bacteriophage and Their Impact on Pseudomonas Virulence, Mammalian Immunity, and Chronic Infections. Front Immunol. 11: 244. https://doi.org/10.3389/fimmu.2020.00244.
- Pei, T. T., Luo, H., Wang, Y., Li, H., Wang, X. Y., Zhang, Y. Q., An, Y., Wu, L. L., Ma, J., Liang, X., et al. (2024). Filamentous prophage Pf4 promotes genetic exchange in Pseudomonas aeruginosa. ISME J. 18(1). https://doi.org/10.1093/ismejo/wrad025.
- Secor, P. R., Sweere, J. M., Michaels, L. A., Malkovskiy, A. V., Lazzareschi, D., Katznelson, E., Rajadas, J., Birnbaum, M. E., Arrigoni, A., Braun, K. R., et al. (2015). Filamentous Bacteriophage Promote Biofilm Assembly and Function. Cell Host Microbe. 18(5): 549–559. https://doi.org/10.1016/j.chom.2015.10.013.
- Tarafder, A. K., von Kugelgen, A., Mellul, A. J., Schulze, U., Aarts, D. and Bharat, T. A. M. (2020). Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria. Proc Natl Acad Sci USA. 117(9): 4724–4731. https://doi.org/10.1073/pnas.1917726117.
- Guerrero-Mandujano, A., Hernandez-Cortez, C., Ibarra, J. A. and Castro-Escarpulli, G. (2017). The outer membrane vesicles: Secretion system type zero. Traffic. 18(7): 425–432. https://doi.org/10.1111/tra.12488.
- Liu, S., Li, Y., Zhang, Y., Seng, Z. J., Xu, H., Yang, L. and Wu, Y. (2022). Self-organized canals enable long-range directed material transport in bacterial communities. eLife. 11. https://doi.org/10.7554/eLife.79780.
- Toyofuku, M., Schild, S., Kaparakis-Liaskos, M. and Eberl, L. (2023). Composition and functions of bacterial membrane vesicles. Nat Rev Microbiol. 21(7): 415–430. https://doi.org/10.1038/s41579-023-00875-5.
- Turnbull, L., Toyofuku, M., Hynen, A. L., Kurosawa, M., Pessi, G., Petty, N. K., Osvath, S. R., Carcamo-Oyarce, G., Gloag, E. S., Shimoni, R., et al. (2016). Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun. 7: 11220. https://doi.org/10.1038/ncomms11220.
- Blackburn, S. A., Shepherd, M. and Robinson, G. K. (2021). Reciprocal Packaging of the Main Structural Proteins of Type 1 Fimbriae and Flagella in the Outer Membrane Vesicles of "Wild Type" Escherichia coli Strains. Front Microbiol. 12: 557455. https://doi.org/10.3389/fmicb.2021.557455.
- Liu, J. H., Zhang, Y., Zhou, N., He, J., Xu, J., Cai, Z., Yang, L., and Liu, Y. (2024). Bacmethy: A novel and convenient tool for investigating bacterial DNA methylation pattern and their transcriptional regulation effects. iMeta. 3(3). https://doi.org/10.1002/imt2.186.
Article Information
Publication history
Received: May 9, 2024
Accepted: Jul 12, 2024
Available online: Aug 2, 2024
Published: Aug 20, 2024
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Li, S., Ren, A., Li, M., Li, G., Yang, L. and Jia, T. (2024). Extraction of Bacterial Membrane Vesicle and Phage Complex by Density Gradient Ultracentrifugation. Bio-protocol 14(16): e5050. DOI: 10.21769/BioProtoc.5050.
Category
Microbiology > Microbial cell biology > Organelle isolation
Microbiology > Microbe-host interactions > Virus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link







