- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparing and Evaluating the Stability of Therapeutically Relevant Oligonucleotide Duplexes
(§ Technical contact) Published: Vol 14, Iss 8, Apr 20, 2024 DOI: 10.21769/BioProtoc.4975 Views: 4019
Reviewed by: Chiara AmbrogioWendy Leanne HempstockCatherine HurdAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Visualization, Quantification, and Modeling of Endogenous RNA Polymerase II Phosphorylation at a Single-copy Gene in Living Cells
Linda S. Forero-Quintero [...] Timothy J. Stasevich
Aug 5, 2022 2485 Views
Profiling of Single-cell-type-specific MicroRNAs in Arabidopsis Roots by Immunoprecipitation of Root Cell-layer-specific GFP-AGO1
Lusheng Fan [...] Xuemei Chen
Dec 20, 2022 1950 Views
Successful Transfection of MicroRNA Mimics or Inhibitors in a Regular Cell Line and in Primary Cells Derived from Patients with Rheumatoid Arthritis
Si Wang [...] Shemin Lu
Sep 20, 2023 1547 Views
Abstract
The field of oligonucleotide therapeutics is rapidly advancing, particularly for combating orphan diseases and cancer. However, the intrinsic instability of oligonucleotides, especially RNA, poses a substantial challenge in the face of the harsh conditions encountered intracellularly and in circulation. Therefore, evaluating the stability of oligos in serum is of great significance when developing oligonucleotide therapeutics. This protocol outlines a dependable and reproducible method for preparing oligonucleotide duplexes, coupled with confirmation by gel electrophoresis. Subsequently, the protocol defines a mechanism to assess the stability of the oligo duplexes in serum. This protocol seeks to establish a standardized reference for researchers, enabling them to compare the impact of various modifications on oligo stability and assess the degradation kinetics effectively.
Key features
• Adaptable for use with small interfering RNA (siRNA), microRNA (miRNA), antisense oligonucleotides (ASOs), and other unmodified and modified oligonucleotides.
• Does not necessitate any Biological Safety Level clearance and offers a rapid, cost-effective, and entirely in vitro procedure.
• Allows researchers to evaluate multiple modification patterns that, when coupled with targeting activity, allow for selecting the best modification pattern prior to in vivo analysis.
Keywords: SerumGraphical overview
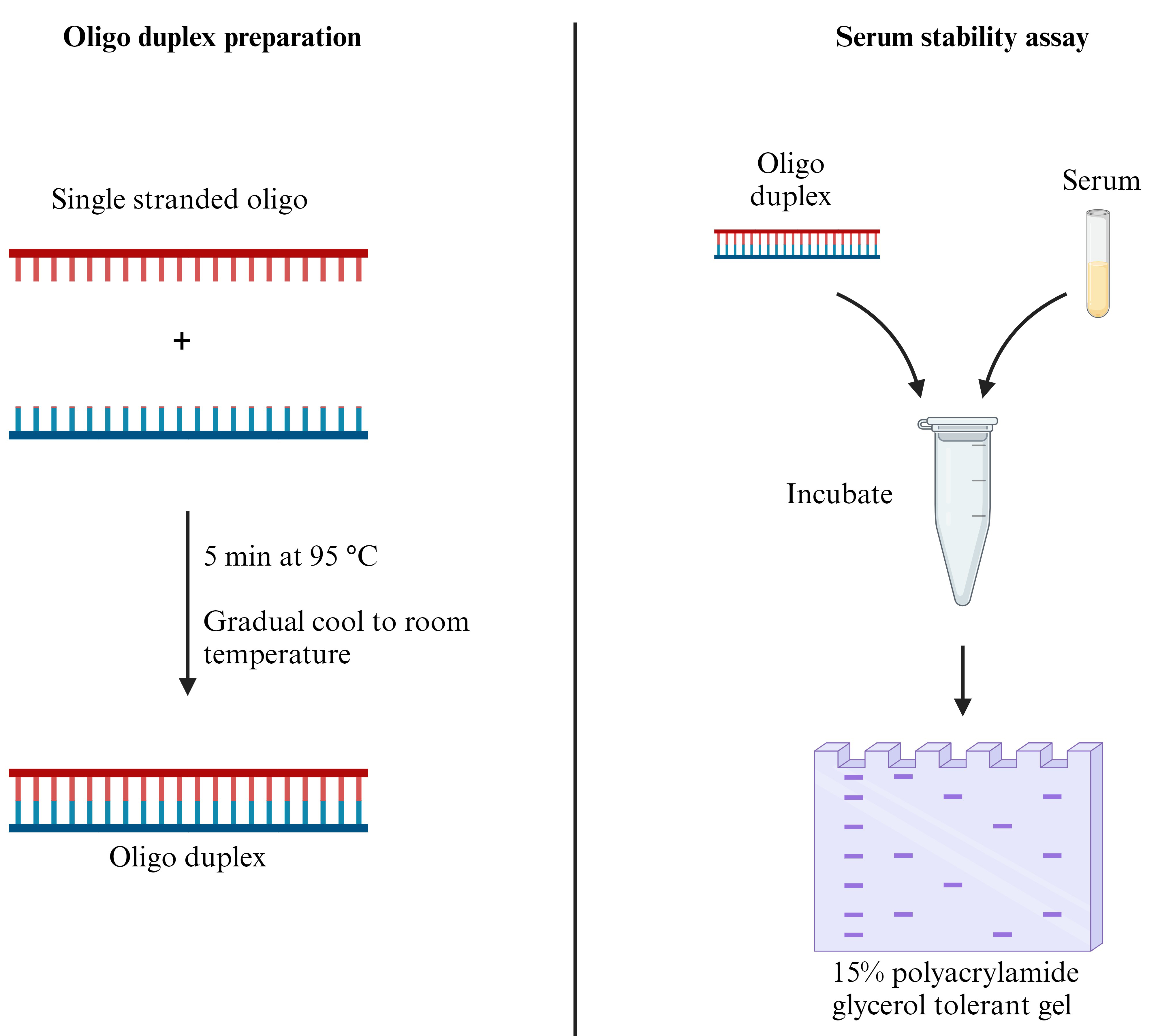
Background
The field of RNA therapeutics holds promise for applications in various diseases. However, the inherent instability of RNA poses a major obstacle, especially due to the harsh conditions encountered during circulation [1]. Therefore, a protocol for rapid in vitro evaluation of RNA stability in serum holds critical significance. This protocol presents a reliable and reproducible method for preparing oligo duplexes followed by the assessment of their stability in serum.
Fetal bovine serum (FBS) is used extensively in research settings as an additive to basal growth medium for cell and tissue culture applications. It serves as a rich source of proteins and growth factors that are vital for supporting cell growth in a cultured environment. FBS contains over 1,000 different components, including growth and attachment factors, lipids, hormones, essential nutrients, energy sources, electrolytes, carriers, and enzymes [2]. Notably, FBS also contains nucleases, which cause degradation of oligonucleotides over time [3,4]. Thus, FBS replicates a surrogate for the conditions faced by oligonucleotides during their circulation in the bloodstream, potentially even more rigorous than those encountered in circulation.
RNA instability, coupled with potential immunogenic effects associated with unmodified RNAs, are critical challenges in the development of RNA-based therapies. Unmodified oligos face rapid degradation by nucleases and elicit immunogenic effects, limiting their efficacy and necessitating high and frequent dosing in vivo [1]. Various chemical modifications, such as 2'-O-methyl and 2'-fluoro modifications to the ribose and phosphorothioate substitutions to the backbone, have been employed to enhance RNA stability [5–7]. Ribose modifications improve binding affinity and nuclease protection, while phosphorothioate bonds confer resistance to exonucleases [8]. The 2'-O-methyl modification also mitigates immune system stimulation triggered by delivered RNA. Although these modifications are advantageous, each RNA requires a unique pattern of modifications along both sense and antisense strands to limit degradation while maintaining target engagement. The protocol provided here allows one to evaluate "the stability" of multiple modification patterns and, when coupled with gene expression analysis (qRT-PCR or western), the researcher can easily identify the optimal modification pattern for a particular RNA.
Due to blood-born nucleases, these RNA modifications are essential, especially when the RNA is delivered using direct ligand conjugation [9–11]. Nonetheless, this protocol's relevance extends to studies involving therapeutic RNA encapsulation and studies using transfection of modified miRNA and siRNA, which are subject to intracellular degradation, effects that can be evaluated using this simple in vitro protocol.
While the concept of serum stability is not novel, the methodologies employed by researchers tend to exhibit variability [12,13]. This protocol aims to serve as a standardized reference for researchers. By utilizing this protocol, researchers can effectively compare the impact of various modifications on the stability of various oligos and evaluate the degradation kinetics of oligonucleotide therapeutics.
Materials and reagents
Biological materials
Fetal bovine serum Premium (Bio-Techne, catalog number: S11150)
Oligonucleotides (Integrated DNA technologies or other relevant source)
Complementary sense and antisense strands
Options to add modified bases including 2'-O-methyl and 2'-fluoro nucleotides, modifications to the backbone, including phosphorothioate backbone modification, replacing phosphodiester bonds in the oligo backbone at specific locations, or other modifications.
Reagents
Nuclease-free water (Invitrogen, catalog number: AM9932)
Tris base, molecular biology grade (Millipore Sigma, catalog number: 648310)
Concentrated hydrochloric acid (HCl) (Fisher Chemical, catalog number: SA49)
Sodium chloride (NaCl) (Millipore Sigma, catalog number: S9888)
EDTA (0.5 M), pH 8.0, RNase-free (Invitrogen, catalog number: AM9260G)
Taurine (Millipore Sigma, catalog number: T0625)
Acrylamide/Bis solution 29:1 (30%) (Bio-Rad, catalog number: 1610156)
Ammonium persulfate (APS) (Millipore Sigma, catalog number: A3678)
TEMED (tetramethylethylenediamine) (Bio-Rad, catalog number: 1610800)
2× RNA loading dye (Thermo Scientific, catalog number: R0641)
GelRed nucleic acid gel stain (Fisher Scientific, Biotium, catalog number: NC9594719)
Phosphate buffered saline (PBS) (Fisher Scientific, catalog number: SH30256FS)
Solutions
NaCl (2.5 M) (see Recipes)
1 M Tris, pH 7.5-8.0 (see Recipes)
10× annealing buffer (see Recipes)
20× glycerol-tolerant gel buffer (see Recipes)
15% polyacrylamide glycerol-tolerant gel (see Recipes)
Recipes
NaCl (2.5 M)
Reagent Final concentration Quantity NaCl (powder) 2.5 M 1.46 g Nuclease-free H2O n/a 10 mL Total 2.5 M 10 mL 1 M Tris, pH 7.5-8.0
Reagent Final concentration Quantity Tris base 1M 121.1 g Nuclease-free H2O n/a 800 mL Concentrated HCl n/a 65 mL Nuclease-free H2O n/a Add to bring final volume to 1 L Total n/a 1 L 10× annealing buffer
Reagent Final concentration Quantity Tris, pH 7.5–8.0 (1 M) (Recipe 2) 100 mM 400 µL NaCl (2.5 M) (Recipe 1) 500 mM 800 µL EDTA (0.5 M) 10 mM 80 µL Nuclease-free H2O n/a 2,720 µL Total n/a 4 mL Store at -20 °C
20× glycerol-tolerant gel buffer
Reagent Final concentration Quantity Tris base 1.78 M 216 g Taurine 570 mM 72 g EDTA (0.5 M) 1 mM 2 mL Nuclease-free H2O n/a Bring to 1,000 mL Total n/a 1,000 mL Store at 4 °C
15% polyacrylamide glycerol-tolerant gel (see Note 4)
Reagent Final concentration Quantity Acrylamide/Bis solution 29:1 (30%) 15% 7.5 mL Glycerol-tolerant gel buffer (20×) (Recipe 4) 1× 750 µL Ammonium persulfate (10%) 0.1% 150 µL TEMED 1% 15 µL Nuclease-free H2O n/a 6.6 mL Total n/a 900 mL
Laboratory supplies
Standard pipette tips with a volume capacity of 2 µL, 20 µL, 200 µL, and 1 mL
Manual pipettes set of 2, 20, 200, and 1,000 µL (Mettler-Toledo, Rainin, catalog number: 17014393, 17014392, 17014391, and 17014382 or similar)
Premium microcentrifuge tubes 1.5 mL (Fisherbrand, catalog number: 05-408-129, or similar RNase-free microcentrifuge tubes)
Bel-Art® gel staining box with cover (Millipore Sigma, catalog number: BAF135511000-1EA)
Equipment
Standard dry block heaters (VWR, catalog number: 75838-318)
Criterion empty cassettes (Bio-Rad, catalog number: 3459901)
Criterion cell and PowerPac basic power supply (Bio-Rad, catalog number: 1656019)
VWR® compact UV transilluminator (VWR, catalog number: 76407-432)
Laboratory shaker (Reliable Scientific, model: 55)
High-resolution digital camera (iPhone, Android, or other cell phone cameras are acceptable)
Software and datasets
ImageJ is a Java-based (runs on Mac OS X, Linux, and Windows) freeware available for download at: http://rsb.info.nih.gov/ij/
Procedure
Oligo duplex preparation
Resuspend each single-stranded oligonucleotide (oligo) strand to a concentration of 200 µM or to the desired concentration using nuclease-free water (Note 1).
Combine 10 µL of each of the 200 µM RNA oligo solutions (sense and antisense) and 5 µL of 10× annealing buffer (Recipe 3) in a 1.5 mL microcentrifuge tube with 25 µL of nuclease-free water. The final volume is 50 µL (Note 2).
Incubate the solution for 5 min at 95 °C in a dry heat block and allow to cool down slowly to room temperature (Critical) (Note 3).
The solution can be stored frozen at -20 °C and freeze-thawed up to five times. The final concentration of oligo duplex is 40 µM and can be diluted to the desired concentration with nuclease-free water (Pause Point).
Analysis of oligo duplexes
Cast a 15% polyacrylamide glycerol-tolerant gel using either 12-well or 18-well combs (Recipe 5) (Note 4).
Place polymerized gels into the gel running apparatus and fill with 1× glycerol-tolerant gel buffer (dilution of 20× glycerol-tolerant gel buffer, Recipe 4).
Gently remove the combs from the gels and clean out the wells by forcefully pipetting the 1× glycerol-tolerant gel buffer into empty wells.
Mix 1 µL of the oligo sample of the desired concentration with 4 µL of water and 5 µL of 2× RNA loading dye (Notes 5 and 6).
Denature RNA/RNA loading dye at 70 °C for 5 min in a dry heat block and transfer samples back to ice quickly.
Load the samples on the gel and run at 100 V until the desired separation is achieved (Note 7).
Once the gel has finished running, carefully separate the glass cassette and remove the gel. Transfer the gel into a small dish for staining.
Stain the gel using GelRed nucleic acid gel stain by mixing 10 µL of GelRed with 10 mL of 1× glycerol-tolerant gel buffer. Incubate the gel in this mixture for 10 min at room temperature with gentle shaking (not to exceed 30% of maximum to avoid gel tear).
Visualize the oligo bands in the gel using a UV transilluminator to confirm successful annealing of the duplex and to assess the purity of samples.
Capture an image using a high-resolution camera (Figure 1). This can be used for quantification of band intensities with ImageJ or similar quantification software.

Figure 1. Polyacrylamide gel image for qualitative assessment of oligo duplex generation. Representative GelRed-stained polyacrylamide gel of a fully modified version of miR-34a (FM-miR-34a) containing 2'-O-methyl and 2'-fluoro bases and phosphorothioate backbone modifications, highlighting successful annealing of oligo duplex as indicated by mobility shifts on the gel. For exact sequence and modification pattern of oligos used for this experiment, refer to Abdelaal et al. [8]. Each lane was loaded with 40 pmol of oligo. A similar quality assessment of additional oligo duplexes can be found in Li et al. [14]; Orellana et al. [9].
Serum stability assay
Prepare 50 pmol of the oligo duplex in 50% FBS in a total volume of 10 µL (Note 8 and 9).
Prepare the correct number of tubes (at room temperature) to reflect the desired timepoints. Recommended timepoints include 0 min, 10 min, 30 min, 1 h, 6 h, 12 h, and 24 h.
Incubate the tubes at 37 °C until the timepoint is reached. At that point, mix 5 µL of the RNA/serum sample with 5 µL of RNA loading dye and store the respective tube at -20 °C (Note 10) (Pause Point).
After the last timepoint, analyze samples on a 15% polyacrylamide gel in glycerol-tolerant gel buffer followed by staining RNA using GelRed nucleic acid gel stain, as outlined in section B (Figure 2).
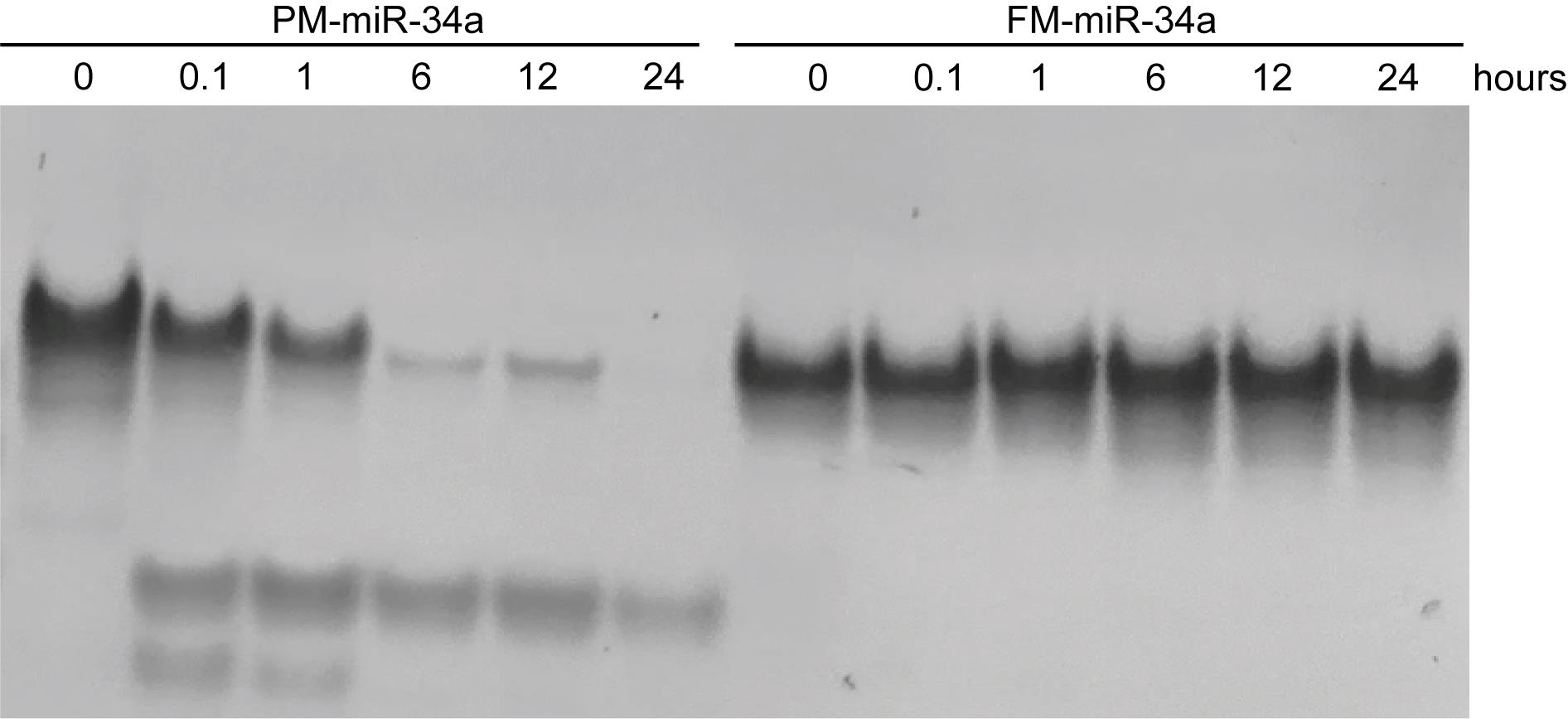
Figure 2. Serum stability gel. Representative GelRed-stained polyacrylamide gel of partially modified (PM) and fully modified (FM) miR-34a following exposure to 50% serum over a time period. For exact sequence and modification pattern of oligos, refer to Abdelaal et al. [8]. Each lane contains 50 pmol of oligo incubated with serum. Figure adapted from Abdelaal et al. [8]. A similar assessment of the stability of duplexes can be found in Li et al. [14]; Orellana et al. [9].
Notes
Throughout the protocol, the stock concentration of the single-stranded oligo is considered to be 200 µM.
The key requirement is a 1:1 equimolar ratio for the two strands, although the concentration of both strands can be higher or lower. The highest concentration tested for annealing is a final concentration of 50 µM for the oligo duplex.
This is accomplished by turning off the heat source but leaving the tube within the dry heat block, allowing it to cool down gradually to room temperature. Alternatively, a beaker of water can be used as a water bath to achieve the 95 °C temperature, and then the heat source is removed, allowing the beaker of water containing the tube of oligos to cool down gradually to room temperature.
This process is optimized for casting gels using Criterion empty cassettes. To avoid premature polymerization, 10% APS and TEMED should be the last two reagents added to the gel solution. After adding TEMED, mix and quickly transfer the appropriate volume of gel solution (~13 mL) to fill up the empty cassette. Insert comb into the gel and allow 20–30 min for the gel polymerization to occur. Do not discard the remainder (~2 mL) of gel solution, as it can be used to verify if the gel solution has completely polymerized. Based on the Criterion gels, gels are typically run at 100 V at room temperature with no concerns of overheating.
If samples were previously frozen, thaw them on ice.
Include necessary controls such as single-stranded oligos and, if desired, a DNA/RNA ladder.
Depending on the size of the oligo, different resolution times may be required to achieve the necessary separation. Run times are typically 90–120 min at room temperature for a microRNA (such as miR-34a), which is 19–25 nucleotides in size per strand.
This is achieved by diluting 1.25 µL of 40 µM duplex by adding 1.25 µL of nuclease-free water and 2.5 µL of 100% FBS. If setting up multiple tubes, a master mix can be created.
The oligo can be incubated with PBS instead of FBS for all timepoints or selected timepoints as negative controls. This ensures that any degradation is attributable to the serum rather than potential nuclease contamination in the sample or water.
For the 0-min timepoint, ensure that the RNA loading dye is added immediately after adding the serum and is frozen immediately.
Data analysis
Measure the band intensities using ImageJ or a similar software. The protocol for using ImageJ can be found in Davarinejad [15].
While one technical replicate is sufficient, at least three biological replicates are necessary due to variability associated with bovine serum.
Student’s t-test can be used to compare band intensities between two separate oligos. ANOVA can be used to compare band intensities across multiple timepoints employed in serum stability assay.
Validation of protocol
This protocol was used to generate and deliver functional microRNA-34a (miR-34a) duplexes, specifically and rapidly to tumor tissues. It was also used to evaluate the development of ligand-conjugated miR-34a [9,11]
This protocol was used to generate and assess the stability of a fully modified miR-34a, compared to the stability of partially modified miR-34a and unmodified miR-34a duplexes following incubation in 50% serum over time. While unmodified and PM-miR-34a were destabilized rapidly following exposure to serum, FM-miR-34a was completely resistant up to 24 h and remained intact for at least 72 h [8].
At least three independent biological replicates are recommended to generate accurate, reliable results.
General notes and troubleshooting
General notes
Originally designed for annealing two complementary synthetic RNA oligos, this protocol has been successfully validated with modified oligos, including 2'-O-methyl and 2'-fluoro ribose sugars, and other backbone modifications including phosphorothioate bonds. It is adaptable for use with small interfering RNA (siRNA), microRNA (miRNA), antisense oligonucleotides (ASOs), and other modified oligos.
This protocol uses FBS for the serum stability assay. It can be modified to include any of the following bovine serum types as necessary: newborn calf serum, bovine calf serum, adult bovine serum, and donor bovine serum.
There is considerable variability between FBS batches, including differing levels of nucleases and other components that contribute to oligo degradation. When using a new batch of FBS, include appropriate controls to enable consistent comparisons with previous results.
For recipes detailed in the Recipes section, prepare the necessary stock solutions first.
The gel employed in this protocol is a polyacrylamide gel run using a glycerol-tolerant buffer. The gel composition has been slightly modified to incorporate glycerol-tolerant buffer within the gel. See Note 4 for additional tips regarding the gel casting process.
Troubleshooting
The final volume while loading might be less than 10 µL—this is likely due to evaporation during RNA denaturation process. This should not affect the outcome of the experiment.
The presence of unannealed single strands visualized in the gel can be corrected by precisely determining the concentration of single-strand products and adding an equal molar concentration during the annealing process.
It is useful to resolve both single-stranded oligos on the gel to gain a better understanding for any lower resolving products on the gel.
Acknowledgments
This work was funded in part by the National Institutes of Health (R01CA205420 and R01CA226259) to A.L.K., and a LCRP Idea Development Award from the Department of Defense (W81XWH-22-1-1085) to A.L.K. This protocol was adapted from Orellana et al. [11], Orellana et al. [9], and Abdelaal et al. [8].
Competing interests
A.L.K. is an inventor on U.S. patent PCT/US2017/061997 submitted by Purdue University that covers methods and uses of ligand-targeted delivery of miRNAs. A.L.K. is an inventor on U.S. provisional patent 63/454,177 submitted by Purdue University that covers methods and uses of modified miR-34a. S.G.I. has no competing interests. A.L.K. is a co-founder and CEO of LigamiR Therapeutics that works on ligand-mediated delivery systems for delivery of microRNAs.
References
- Abdelaal, A. M. and Kasinski, A. L. (2021). Ligand-mediated delivery of RNAi-based therapeutics for the treatment of oncological diseases. NAR Cancer 3(3): e1093/narcan/zcab030.
- Boone, C. W., Mantel, N., Caruso, T. D., Kazam, E. and Stevenson, R. E. (1971). Quality control studies on fetal bovine serum used in tissue culture. In Vitro 7(3): 174–189.
- Eder, P. S., DeVINE, R. J., Dagle, J. M. and Walder, J. A. (1991). Substrate Specificity and Kinetics of Degradation of Antisense Oligonucleotides by a 3′ Exonuclease in Plasma. Antisense Res. Dev. 1(2): 141–151.
- Yen, A., Cheng, Y., Sylvestre, M., Gustafson, H. H., Puri, S. and Pun, S. H. (2018). Serum Nuclease Susceptibility of mRNA Cargo in Condensed Polyplexes. Mol. Pharmaceutics 15(6): 2268–2276.
- Biscans, A., Caiazzi, J., Davis, S., McHugh, N., Sousa, J. and Khvorova, A. (2020). The chemical structure and phosphorothioate content of hydrophobically modified siRNAs impact extrahepatic distribution and efficacy. Nucleic Acids Res. 48(14): 7665–7680.
- Hassler, M. R., Turanov, A. A., Alterman, J. F., Haraszti, R. A., Coles, A. H., Osborn, M. F., Echeverria, D., Nikan, M., Salomon, W. E., Roux, L., et al. (2018). Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res. 46(5): 2185–2196.
- Jackson, A. L., Burchard, J., Leake, D., Reynolds, A., Schelter, J., Guo, J., Johnson, J. M., Lim, L., Karpilow, J., Nichols, K., et al. (2006). Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12(7): 1197–1205.
- Abdelaal, A. M., Sohal, I. S., Iyer, S., Sudarshan, K., Kothandaraman, H., Lanman, N. A., Low, P. S. and Kasinski, A. L. (2023). A first-in-class fully modified version of miR-34a with outstanding stability, activity, and anti-tumor efficacy. Oncogene 42(40): 2985–2999.
- Orellana, E. A., Abdelaal, A. M., Rangasamy, L., Tenneti, S., Myoung, S., Low, P. S. and Kasinski, A. L. (2019). Enhancing MicroRNA Activity through Increased Endosomal Release Mediated by Nigericin. Mol. Ther. Nucleic Acids 16: 505–518.
- Rangasamy, L., Chelvam, V., Kanduluru, A. K., Srinivasarao, M., Bandara, N. A., You, F., Orellana, E. A., Kasinski, A. L. and Low, P. S. (2018). New Mechanism for Release of Endosomal Contents: Osmotic Lysis via Nigericin-Mediated K+/H+ Exchange. Bioconjugate Chem. 29(4): 1047–1059.
- Orellana, E. A., Tenneti, S., Rangasamy, L., Lyle, L. T., Low, P. S. and Kasinski, A. L. (2017). FolamiRs: Ligand-targeted, vehicle-free delivery of microRNAs for the treatment of cancer. Sci. Transl. Med. 9(401): eaam9327.
- Jafari, M., Xu, W., Pan, R., Sweeting, C. M., Karunaratne, D. N. and Chen, P. (2014). Serum Stability and Physicochemical Characterization of a Novel Amphipathic Peptide C6M1 for SiRNA Delivery. PLoS One 9(5): e97797.
- Kajino, R., Sakamoto, S. and Ueno, Y. (2022). Synthesis, gene silencing activity, thermal stability, and serum stability of siRNA containing four (S)-5′-C-aminopropyl-2′-O-methylnucleosides (A, adenosine; U, uridine; G, guanosine; and C, cytidine). RSC Adv. 12(18): 11454–11476.
- Li, W., Wang, Y., Liu, X., Wu, S., Wang, M., Turowski, S. G., Spernyak, J. A., Tracz, A., Abdelaal, A. M., Sudarshan, K., et al. (2024). Developing Folate-Conjugated miR-34a Therapeutic for Prostate Cancer: Challenges and Promises. Int. J. Mol. Sci. 25(4): 2123.
- Davarinejad, H. (2015). Quantifications of Western Blots with ImageJ [Protocol]. https://www.yorku.ca/yisheng/Internal/Protocols/ImageJ.pdf
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Iyer, S. G. and Kasinski, A. L. (2024). Preparing and Evaluating the Stability of Therapeutically Relevant Oligonucleotide Duplexes. Bio-protocol 14(8): e4975. DOI: 10.21769/BioProtoc.4975.
Category
Biochemistry > RNA > Single-molecule Activity
Molecular Biology > RNA > miRNA interference
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link








