- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Published: Vol 14, Iss 7, Apr 5, 2024 DOI: 10.21769/BioProtoc.4970 Views: 2098
Reviewed by: Kristin L. ShinglerEmmanuel Orta-ZavalzaAna Martinez

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Thylakoid Membranes from the Cyanobacterium Synechocystis sp. PCC 6803 and Analysis of Their Photosynthetic Pigment-protein Complexes by Clear Native-PAGE
Josef Komenda [...] Tomas Zakar
Jan 5, 2019 8597 Views

From Llama to Nanobody: A Streamlined Workflow for the Generation of Functionalised VHHs
Lauren E.-A. Eyssen [...] Raymond J. Owens
Mar 20, 2024 6250 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2171 Views
Abstract
Periodontal disease is characterized by the destruction of the hard and soft tissues comprising the periodontium. This destruction translates to a degradation of the extracellular matrices (ECM), mediated by bacterial proteases, host-derived matrix metalloproteinases (MMPs), and other proteases released by host tissues and immune cells. Bacterial pathogens interact with host tissue, triggering adverse cellular functions, including a heightened immune response, tissue destruction, and tissue migration. The oral spirochete Treponema denticola is highly associated with periodontal disease. Dentilisin, a T. denticola outer membrane protein complex, contributes to the chronic activation of pro-MMP-2 in periodontal ligament (PDL) cells and triggers increased expression levels of activators and effectors of active MMP-2 in PDL cells. Despite these advances, no mechanism for dentilisin-induced MMP-2 activation or PDL cytopathic behaviors leading to disease is known. Here, we describe a method for purification of large amounts of the dentilisin protease complex from T. denticola and demonstrate its ability to activate MMP-2, a key regulator of periodontal tissue homeostasis. The T. denticola dentilisin and MMP-2 activation model presented here may provide new insights into the dentilisin protein and identify potential therapeutic targets for further research.
Key features
• This protocol builds upon a method described by Cunningham et al. [1] for selective release of Treponema outer membrane proteins.
• We adapted the protocol for the purification of biologically active, detergent-stable outer membrane protein complexes from large batch cultures of T. denticola.
• The protocol involves large-scale preparative electrophoresis using a Model 491 Prep Cell.
• We then use gelatin zymography to demonstrate the activity of the purified dentilisin complex by its ability to activate matrix metalloproteinase 2 (MMP-2).
Keywords: Treponema denticolaBackground
Periodontitis is a chronic inflammatory disease resulting from bacterial dysbiosis and unfavorable host–bacterial interactions. This ultimately leads to the destruction of periodontal tissues. The development of dysbiotic oral biofilms is associated with the emergence of periodontal disease. This condition is often attributed to elevated levels of the "red complex" bacteria, which include Treponema denticola, Porphyromonas gingivalis, and Tannerella forsythia [2], while several other periodontopathogenic species have been identified in recent studies [3–5]. Previous studies using 16S rRNA and shotgun sequencing have established notable dissimilarities in the microbial communities of individuals with good oral health vs. those with periodontitis. Periodontitis is a complex condition that is not solely caused by one type of microbe. Instead, it is thought to be influenced by various factors, such as changes in inflammation and interactions between the host and microbes, particularly when keystone pathogens colonize [6]. Also, it is important to highlight that various factors play crucial roles in the development of periodontitis, as they can either support or hinder the equilibrium of oral microorganisms.
In this study, we focused on the contribution of the most well-known and well-characterized periodontal pathogenic oral spirochete, namely T. denticola, which is highly associated with periodontal disease. However, T. denticola is often below detectable levels in healthy gingival plaque [7,8]. T. denticola, the most readily cultivable oral spirochete, is the model organism for studying spirochete–host interactions in periodontitis. The severity of periodontitis is directly linked to an increase in the colonization levels of T. denticola, highlighting its significant contribution to the disease [9,10]. One of the factors contributing to the virulence of T. denticola is its protease complex called dentilisin. Dentilisin is an outer membrane–associated complex consisting of the acylated subtilisin-family PrtP protease and two other lipoproteins (PrcB and PrcA) that are unique to oral spirochetes (reviewed in [11]). Dentilisin is encoded as an operon in the chromosome consisting of prcB-prcA-prtP. Dentilisin proteolytic activity directly affects host cells and tissue: it disrupts intercellular junctions [12], contributes to tissue penetration [13], specifically cleaves host proteins critical to maintaining tissue homeostasis [14], and induces dysregulation of TLR/MyD88 and integrin/FAK signaling mechanisms [15–17]. The multifaceted action of T. denticola dentilisin not only facilitates T. denticola's adhesion to and harm of epithelial cells and fibroblasts but also aids in penetrating epithelial tissue; furthermore, it may be involved in T. denticola's strategies to evade complement-mediated bactericidal activity [12,18–20]. One of the unique features of the dentilisin complex is that it retains its stability and proteolytic activity when unheated samples are subjected to SDS-PAGE. We and others have exploited this feature by using electrophoretic methods to purify dentilisin and characterize its activity [14,21,22]. We have previously reported the use of preparative continuous polyacrylamide gel electrophoresis (PC-PAGE) to purify several T. denticola outer membrane–associated proteins [20,22–24]. Here, we describe a method for purifying large amounts of the active dentilisin complex using this method.
The present study focuses on exploring the link between dentilisin and MMP-2 activation in relation to periodontitis. Here, we describe a methodology for the purification of substantial amounts of the dentilisin protease complex from T. denticola and demonstrate its capacity to activate MMP-2, a key regulator of periodontal tissue homeostasis. We present the protocols used in this study in three sections: I. Detergent extraction of T. denticola outer membrane components; II. PC-PAGE for purification of T. denticola dentilisin; and III. Gelatin zymography to detect activation of MMP-2 by dentilisin.
Part I: Detergent extraction of T. denticola outer membrane components
Materials and reagents
Biological materials
Treponema denticola MHE strain [25]. This strain (derived from T. denticola ATCC 35405) is an isogenic mutant that lacks Msp, a prominent outer membrane protein. We used T. denticola MHE for the present study because it is difficult to separate dentilisin from Msp using the wild-type ATCC 35405 strain [20,24]. This strain is available from the authors upon request. While the mutation carried in T. denticola MHE was selected by resistance to erythromycin, antibiotic pressure is not required to maintain the mutation.
Reagents
Tryptone (Fisher Scientific, catalog number: 211705)
Brain heart infusion (Fisher Scientific, catalog number: 237500)
Yeast extract (Fisher Scientific, catalog number: 611805000)
Gelatin (Bio-Rad Laboratories, catalog number: 1706537)
(NH4)2SO4 (Thermo Scientific, catalog number: OXCM0635B)
MgSO4·7H2O (Thermo Scientific, catalog number: AAA144910I)
K2HPO4 (Fisher Scientific, catalog number: BP363)
KH2PO4 (Fisher Scientific, catalog number: BP362)
NaCl (Fisher Scientific, catalog number: S271)
KOH (Fisher Scientific, catalog number: P250)
KCl (Fisher Scientific, catalog number: BP366)
Na2HPO4 (Sigma Millipore, catalog number: 567547)
MgCl2 (Fisher Scientific, catalog number: BP214)
Thiamine pyrophosphate (TPP) (Sigma-Aldrich, catalog number: C8754)
Glucose (Sigma-Aldrich, catalog number: G5400)
L-cysteine HCl (Fisher Scientific, catalog number: BP376)
Sodium pyruvate (Fisher Scientific, catalog number: BP356)
Glacial acetic acid (Fisher Scientific, catalog number: A38S)
Propionic acid (Sigma-Aldrich, catalog number: P1386)
N-butyric acid (Sigma-Aldrich, catalog number: B10355-0)
N-valeric acid (Sigma-Aldrich, catalog number: V9759)
Isobutyric acid (Sigma-Aldrich, catalog number: I1754)
Isovaleric acid (Sigma-Aldrich, catalog number: 12954-2)
D,L-methylbutyric acid (Sigma-Aldrich, catalog number: 193070)
Rabbit serum*, heat inactivated (Pel Freeze Biologicals, catalog number: 31127)
Triton X-114 (Anapoe-X-114) (Anatrace, catalog number: APX114)
Acetone (Fisher Scientific, catalog number: A18)
Tris base (Fisher Scientific, catalog number: BP152)
HCl (12.1 N) (Fisher Scientific, catalog number: A144)
1 M Tris pH 6.8 (Fisher Scientific, catalog number: BP152)
DTT (Fisher Scientific, catalog number: BP172)
SDS (Fisher Scientific, catalog number: BP166)
Glycerol (Fisher Scientific, catalog number: BP229)
Bromophenol blue (BPB) (Fisher Scientific, catalog number: BP114)
Solutions
TYGVS growth medium and supplements for T. denticola (see Recipes)
1 M Tris, pH 8 (see Recipes)
Bromophenol blue (BPB) gel loading solution (see Recipes)
Phosphate buffered saline containing 5 mM MgCl2 (PBS) (see Recipes)
Recipes
TYGVS growth medium and supplements for T. denticola [26,27]
TYGVS base growth medium
Add to approximately 900 mL of distilled water:
10.0 g of tryptone
5.0 g of brain heart infusion
10.0 g of yeast extract
10.0 g of gelatin
0.5 g of (NH4)2SO4
0.1g of MgSO4·7H2O
1.13 of K2HPO4
0.9 g of KH2PO4
1.0 g of NaCl
pH to 7.2 with 4 N KOH in distilled H2O
Autoclave (121 °C, 20 min, 15 psi). Unsupplemented media can be stored for up to six months at 4 °C.
Add 100 mL of TYGVS supplements (described below) aseptically before use.
Complete media with supplements can be stored for up to one month at 4 °C.
TYGVS supplements
Combine the following together, adjust to pH 7.2 with 4 N KOH, and filter sterilize.
Amount per liter of final volume of medium:
0.0125 g of TPP
1.0 g of glucose
1.0 g of L-cysteine HCl
0.25 g of sodium pyruvate
0.27 mL of glacial acetic acid
0.1 mL of propionic acid
0.064 mL of n-butyric acid
0.016 mL of n-valeric acid
0.016 mL of isobutyric acid
0.016 mL of isovaleric acid
0.016 mL of D,L-methylbutyric acid
100 mL of rabbit serum*, heat inactivated
Mix well; pH to 7.2 with 4 N KOH.
Sterilize using a 0.2 μm vacuum filter unit.
*Note: Heat-inactivated horse or bovine serum can also be used, with little difference in growth.
1 M Tris, pH 8
Mix 12.11 g of Tris in 80 mL of H2O. Adjust to pH 8 by dropwise addition of 1M HCl.
Bromophenol blue (BPB) gel loading solution (10 mL)
Reagent Final concentration Quantity or Volume 1 M Tris pH 6.8 in distilled H2O 360 mM 3.75 mL DTT 600 mM 0.93 g SDS 12% 1.2 g Glycerol 60% 6 mL BPB 0.018% 18 mg Phosphate buffered saline containing 5 mM MgCl2 (PBS)
NaCl 80 g/L
KCl 2 g/L
Na2HPO4 14.4 g/L
KH2PO4 2.4 g/L
MgCl2 1.0 g/L
Distilled H2O, 900 mL
pH to 7.4 with 4 N KOH; then, correct volume to 1 L with H2O
Laboratory supplies
Culture growth vessels. Size is limited by the ability to pass through anaerobic chamber port. We typically use 2 L Corning Pyrex bottles (Corning, catalog number: 13952L)
Spectrophotometry cuvettes: Polystyrene, 2 mL (Fisher Scientific, catalog number: 14955127)
Centrifuge tubes:
250 mL (nominal), Nalgene, polypropylene copolymer (Thermo Scientific, catalog number: 31410250). These tubes will fit a GSA-type rotor
50 mL (nominal), Oak Ridge style (Thermo Scientific, catalog number: 31190050) (polypropylene copolymer) and 31180050 (polycarbonate). These tubes will fit a SS-34-type rotor
15 mL, conical, polypropylene (Corning, catalog number: 430766 or equivalent). These tubes will fit the appropriate swinging bucket rotor insert in STR40R or equivalent centrifuge
50 mL, conical, polypropylene (Corning, catalog number: 430291) or equivalent. These tubes will fit the appropriate swinging bucket rotor insert in STR40R or equivalent centrifuge
Vacuum filter units (0.2 µm) (Thermo Scientific, catalog number: 5650020)
Equipment
Anaerobic chamber (Coy Lab Products or equivalent). We use the Coy Type B vinyl anaerobic chamber. Coy and other manufacturers also make rigid (metal- or polymer-walled) anaerobic chambers, either with gloves or gloveless, that are advertised as having equivalent performance to the “classic” vinyl chamber
Compressed gas tanks: Pre-purified nitrogen (NITPP; Cryogenic Gases, Detroit, MI); mixed gas (10% H2, 5% CO2, 85% N2) (5CO10H2N2; Cryogenic Gases, Detroit, MI). Specialty gas tank regulators (Grainger, Lake. Forest, IL)
Class II biological safety cabinet (Advance SG403, Baker, Sanford, ME). T. denticola is generally classified as an RG2 microbe requiring BSL2 containment and procedures
High-speed centrifuge with rotors for 15 mL, 50 mL, and 250 mL tubes
Sorvall RC2-B (or equivalent) with GSA rotor for 250 mL tubes; SS-34 rotor for 50 mL Oak Ridge tubes
Sorvall ST40R (or equivalent) equipped with swinging bucket rotor for 15 mL and 50 mL tubes
Vacuum trap system for biohazardous waste
SPE vacuum pump trap kit (Millipore Sigma, catalog number: 57120-U)
Whatman Vacu-Guard in-line filter (Cytiva, catalog number: 67225001)
Laboratory “house” vacuum system or small vacuum pump (Southern Labware, catalog number: 167300-11)
pH meter: Accumet AE 150 (or equivalent) (Fisher Scientific, catalog number: 13-636-AE153)
Spectrophotometer (Dayton, Model 1200, Unico)
Platform shaker (Labnet Orbit 1000, catalog number: S20301000B)
Procedure
Detergent extraction and phase partitioning of T. denticola for separation of outer membrane proteins. This procedure gently solubilizes the T. denticola outer membrane, releasing outer membrane and periplasmic proteins while leaving the protoplasmic cylinder intact. A solution of the nonionic detergent Triton-X114 is homogeneous at 0 °C but separates into an aqueous phase and a detergent phase above 20 °C [28–30]. This feature can be utilized to greatly enrich hydrophobic membrane-associated lipoproteins in the detergent phase while periplasmic proteins segregate in the aqueous phase. This methodology is used in spirochete research, both to isolate their abundant lipoproteins and to distinguish between soluble periplasmic proteins and outer membrane components [1,14,20,30–33]. While many investigators incorporate a protease inhibitor cocktail in this procedure, we do not include this step here because it would preclude our objective of obtaining a proteolytically active protein complex. The steps involved in this procedure for detergent extraction and preparation of samples for preparative electrophoresis are illustrated in Figure 1.
Grow T. denticola strain MHE [25] at 37 °C under anaerobic conditions (10% H2, 5% CO2, 85% N2) in complete TYGVS medium [26,27] to late log phase (3–4 days). The expected optical density at 600 nm is approximately 0.3–0.4 as measured in a standard spectrophotometer. The MHE strain is used for dentilisin purification because it lacks the major outer membrane protein Msp, which is difficult to separate from dentilisin using these methods. We typically grow up to 4 L of culture (up to 2 L per culture bottle) for this procedure, which will yield approximately 1.5 mL of the final step extract per liter of culture at a concentration of approximately 500 µg/mL [14].
Harvest culture by centrifugation (4,000× g, 10 min, 4 °C) using 250 mL Nalgene centrifuge tubes. Resuspend in half of the original culture volume of PBS (pH 7.4) containing 5 mM MgCl2, centrifuge as above, and repeat the wash/centrifugation step twice. At each step, suspend the pellet by pipetting gently in a small volume (10–20 mL) of the same PBS+MgCL2 buffer solution and then adding the remaining volume.
Resuspend the pellet as above in 1/40 of the original culture volume of PBS containing 5 mM MgCl2 and 1% Triton X-114.
Note: Triton X-114 is supplied as a 10% aqueous solution. Use small-volume containers of Triton X-114 because it easily becomes oxidized after exposure to air.
Rock the suspension gently overnight at 4 °C in Oak Ridge tubes on a platform shaker.
Centrifuge at 20,000× g for 10 min at 4 °C to separate and pellet unsolubilized material, which is primarily protoplasmic cylinders of spirochete cells [35].
Transfer the clear supernatant containing detergent-soluble material to new 15 mL conical tubes. Adding a drop of bromophenol blue solution (0.04%) to the mixture facilitates visualization of the detergent phase in the following separation step without affecting proteins of interest [32]. The extract may be processed immediately or stored at -80 °C until further processing, which is generally done within a few days.
Incubate the extract at 37 °C for 30 min. The solution will become cloudy.
Prewarm a rotor to 37 °C and then centrifuge the extract at 3,500× g for 10 min at 37 °C in a swinging bucket rotor. The extract should separate into an aqueous upper phase and a detergent lower phase of approximately 1/10 total volume. If good separation is not apparent, centrifugation speed and time can be increased to as much as 20,000× g for 1 h [1,30]. If this is necessary, perform centrifugation in polycarbonate Oak Ridge tubes, because they tolerate higher speeds and allow easier visualization of the detergent pellet than similar polypropylene tubes.
Carefully remove all traces of the aqueous phase from the lower phase.
Dilute the remaining detergent phase with PBS containing 5 mM MgCl2 (4 °C) to bring to the original volume before phase separation. This will return the Triton X-114 concentration to approximately 1%. Repeat phase separation steps 4–9 twice.
Transfer the final lower (detergent) phase to a fresh 50 mL polypropylene Oak Ridge tube. Note that polycarbonate tubes are incompatible with acetone.
Precipitate the detergent phase with acetone to remove Triton X-114 detergent from the sample prior to electrophoresis. Add 8 volumes of cold acetone (-20 °C) and incubate at -20 °C for at least 1 h but not more than 20 h. Centrifuge at 12,000× g at 4 °C for 30 min. Carefully pour off acetone from the soft pellet. Allow the pellet to partially dry and then dissolve in 1/2 volume of PBS containing 5 mM MgCl2. Detergent phase samples generally do not require further concentration before PC-PAGE.
The sample may be stored at -80 °C until purification on Prep Cell, as described in the following section. We generally try to proceed to the PC-PAGE step as soon as is practicable.
Part II: PC-PAGE for purification of T. denticola dentilisin complex
The dentilisin protease complex can be purified from the detergent phase extract prepared in the prior steps by preparative electrophoresis in a Model 491 Prep Cell. The complex, consisting of three lipoproteins (PrcB, PrcA, and PrtP) is remarkably stable under SDS-PAGE conditions when not heated prior to electrophoresis and retains proteolytic activity [13,21,24,36,37]. This PC-PAGE technique is appropriate for the purification of proteins whose apparent molecular weight differs by greater than 5% from their nearest neighbors in SDS-PAGE. In the case of dentilisin, the detergent extraction/phase-partitioning procedure removes the bulk of the proteins from the original crude preparation, and the approximately 95 kDa native protease complex can be recovered efficiently at a level of purity appropriate for many functional assays.
Pre-run preparation is done the day prior to starting the gel run. Including the pre-run, the whole process through sample collection takes approximately three days, not including characterization of fractions by SDS-PAGE and silver staining.
Materials and reagents
Biological materials
Detergent phase extract of T. denticola MHE prepared above
Reagents
30% Acrylamide/Bis solution, 37.5:1 (Bio-Rad Laboratories, catalog number: 161-0158)
Tris pH 8.8, 1.5 M in H2O
Tris-HCl pH 6.8, 1.25 M in H2O
Glycine (Fisher Scientific, catalog number: BP381)
Sodium dodecyl sulfate (SDS) (Invitrogen, catalog number: 15525017)
Tetramethylethylenediamine (TEMED) (Invitrogen, catalog number: 15524)
Ammonium persulfate (APS) (Thermo Scientific, catalog number: 32708)
Bromophenol blue 6× dye solution (Thermo Scientific, catalog number: J61337.AC)
CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate) (Sigma-Aldrich, catalog number: C3023)
Bicinchoninic acid (BCA) protein assay kit (Thermo Scientific catalog number: 23225)
Horseradish peroxidase (HRP)-conjugated Goat Anti-Rabbit IgG (H+L) (Bio-Rad Laboratories, catalog number: 1706515)
SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, catalog number: 34580)
Dentilisin activity detection:
N-Succinyl-L-alanyl-L-alanyl-L-prolyl-L-phenylalanine-p-nitroanilide (SAAPFNA) (Sigma-Aldrich, catalog number: S7388)
N,N-dimethyl formamide (Thermo Scientific, catalog number: AC42364)
SAAPFNA stock: 50 mM in N,N-dimethyl formamide. Store at -20 °C
Reaction buffer: (1×) 50 mM Tris, 0.2 M NaCl, 5 mM DTT, pH 7.5. Make as 10× stock and store at -20 °C.
α-chymotrypsin, Type II as control. Dilute in reaction buffer before use: 1 mg/mL and further 1/10 dilutions (Sigma-Aldrich, catalog number: C4129)
Flat-bottom 96-well microplates (Fisher Scientific, catalog number: FB012931)
Solutions
6× Laemmli SDS sample buffer (see Recipes)
Upper electrophoresis chamber (cathode) buffer (see Recipes)
Lower electrophoresis chamber (anode) buffer (see Recipes)
Lower electrophoresis chamber (anode) buffer (see Recipes)
Elution buffer (see Recipes)
Post-electrophoresis buffer exchange (see Recipes)
APS 10% in H2O (freshly made)
Recipes
Standard SDS-PAGE preparative gel column
Resolving gel: 7.5% acrylamide for 28 mm diameter column with a gel height of 9.5 cm.
Scale appropriately for 37 mm column or for different acrylamide concentrations.
Note: Acrylamide monomer is a neurotoxin.
30% acrylamide/Bis 37.5:1 9.5 mL
1.5 M Tris pH 8.8 9.5 mL
H2O 18.4 mL
10% SDS 0.38 mL
TEMED 0.02 mL
10% APS 0.2 mL
Stacking gel: 2× sample volume (assuming 5 mL sample volume)
30% acrylamide/Bis 37.5:1 1.3 mL
1.25 M Tris-HCl pH 6.8 1.0 mL
H2O 7.54 mL
10% SDS 0.10 mL
TEMED 0.01 mL
10% APS 0.05 mL
6× Laemmli SDS sample buffer
375 mM Tris-HCl/Tris Base
9% SDS (w/v)
50% Glycerol (v/v)
9% 2-mercaptoethanol (v/v)
0.075% Bromophenol blue (w/v)
Upper electrophoresis chamber (cathode) buffer (500 mL)
25 mM Tris (pH 8.8)
192 mM glycine
0.1% SDS
Lower electrophoresis chamber (anode) buffer (2,000 mL)
25 mM Tris (pH 8.8)
192 mM glycine
Elution buffer (700 mL)
25 mM Tris (pH 8.8)
192 mM glycine
Notes:
We omit SDS from the anode and elution buffers so that there will be minimal residual detergent in the fractions collected. We have seen no effect of this modification on electrophoresis and fraction collection.
Degas all buffers and frits before use. Put the frits in a dish and cover with Tris/glycine buffer. Place them in the port of the anaerobic chamber with caps loosened, pull a vacuum, incubate overnight, then tighten caps, and hold at 4 °C until use.
Post-electrophoresis buffer exchange
PBS containing 0.1% CHAPS
Equipment
Chromatography refrigerator or cold room (Thermo Scientific, TSX2305CA)
Model 491 Prep Cell (Bio-Rad Laboratories, catalog number: 1702928)
Power supply (Bio-Rad Laboratories, model: PowerPac 3000)
Low-pressure chromatography system (Bio-Rad Laboratories, model: BioLogic LP)
ChemiDoc MP image system (Bio-Rad, model: Universal hood III, catalog number: 731BR01488)
Model 2128 fraction collector (flow rate: 1 mL/min; fractions: 5.0 mL; up to two racks of 128 tubes each) (Bio-Rad Laboratories, catalog number: 731-8123)
Variable speed buffer recirculation pump (Bio-Rad Laboratories, catalog number: 1702929)
Sample concentrators:
Millipore/Amicon stirred ultrafiltration cell. Several sizes are available: 50 mL (Millipore, catalog number: UFSC05001), 200 mL (Millipore, catalog number: UFSC20001), and 400 mL (Millipore, catalog number: UFSC40001). This is a stand-alone apparatus that requires a dedicated nitrogen gas tank to supply filtration pressure. Use with ultrafiltration disc membranes designed for use with stirred cells. Disc sizes are available for each size of stirred cell (Cole-Parmer, Inc., model: Millipore EW-29949)
Amicon Ultracel 30 centrifugal concentration unit (Millipore, catalog number: UFC9030). These are single-use filtration units appropriate for use in an ST40R centrifuge. They are available in a wide range of other volumes and molecular weight cutoffs. While relatively inexpensive, costs add up rapidly on large projects
Compressed nitrogen gas tank (NITPP; Cryogenic Gases, Detroit, MI)
Specialty gas regulator for nitrogen tank (Grainger, model: Harris KH1008). Stirred cell concentrators utilize gas pressure for sample concentration
Microplate reader (Molecular Devices, model: VERSAmax). A plate reader is used to assay dentilisin activity against the chromogenic substrate N-succinyl-L-alanyl-L-alanyl-L-prolyl-L-phenylalanine-p-nitroanilide (SAAPFNA) [21]
Procedure
Detailed instructions for assembly and operation of the Model 491 Prep Cell are provided in the Bio-Rad instruction manual. The steps involved in preparative electrophoresis of the sample prepared in Part I above are illustrated in Figure 1. The following are specific items for this application.
Assemble the 28 mm diameter glass gel tube assembly on its casting stand as described in the manual. For larger batch preps, it may be useful to use the 37 mm diameter glass gel tube, with gel component volumes scaled up accordingly.
Attach a continuous flow of water on ice through the cooling core after setting up on the casting stand.
Prepare 7.5% acrylamide resolving gel (total volume: 38 mL): 9.5 mL of 30% acrylamide/Bis 37.5:1; 9.5 mL of 1.5 M Tris pH 8.8; 16.55 mL of dH2O; 0.38 mL of 10% SDS; 0.038 mL of TEMED; 2.66 mL of 1% APS.
Pour resolving gel, one-third of the volume at a time, and then tap and shake to ensure there are no bubbles at the bottom or the side of the column each time. Top with 2 mL of H2O-saturated butanol.
After ~1 h polymerization, discard butanol, wash twice with distilled H2O, and cover with 25 mM Tris (pH 8.8), 192 mM glycine, and 0.1% SDS. This can be left for 48 h or longer at 4 °C or overnight at room temperature (RT) to ensure that polymerization is complete.
Pour an appropriate volume of stacking gel (= 2× sample volume). Note that it is not uncommon to load a 5 mL sample on the Prep Cell. For 10 mL of stacking gel: 7.66 mL of H2O, 1.0 mL of 1.25 M Tris-HCl pH 6.8; 0.1 mL of 10% SDS; 0.01 mL of TEMED; 0.07. mL of 1% APS. If the resolving gel is at RT, allow the stacking gel to polymerize for 30 min.
Disassemble the casting stand and assemble the gel column in the Model 491 Prep Cell according to the manufacturer's instructions, with particular attention to proper placement of support frit, elution frit, dialysis membrane, and O-rings.
In the chromatography refrigerator (or cold room), connect each of the following according to the instructions in the BioLogic LP chromatography manual:
Buffer cooling core inlet from the upper buffer chamber.
Buffer cooling core outlet into the buffer circulation pump.
Outlet from the pump to the spigot of the lower buffer chamber.
Elution outlet at the middle of the top of the cooling core to the inlet of BioLogic System pump.
Be sure you have a separate collection container for the elution buffer that comes off before initiation of fraction collecting.
Add 25 mM Tris (pH 8.8) containing 192 mM glycine to the lower buffer chamber above the level of the stacking gel.
Thaw the detergent phase extract on ice and add 1/5 volume standard 6× SDS-PAGE sample buffer containing reducing agent.
Add the sample with a Pasteur pipette via cut off plastic pipette guide as described in the Prep Cell Instruction Manual. It is useful to become familiar with this gel-loading procedure ahead of time. After finishing, remove the plastic pipette guide by sealing the top opening of the guide with your finger and gently pulling the guide out.
Fill the upper buffer chamber with 25 mM Tris (pH 8.8), 192 mM glycine, and 0.1% SDS. Fill the elution buffer chamber with 25 mM Tris (pH 8.8) and 192 mM glycine buffer.
Connect the electrodes of the Prep Cell to the Bio-Rad PowerPac. Set at 60 mA constant current, which should result in an initial voltage of approximately 550 V.
Connect the buffer circulation pump with 1.6 mm tubing and set the flow rate to 1 mL/min.
Do not turn on the fraction collector until needed (i.e., until after the dye front has passed through the gel). Set fraction collection to collect 5 mL fractions at a flow rate of 1 mL/min.
Screen every tenth fraction by silver-stained SDS-PAGE (10 µL per lane). Pool fractions containing the expected dentilisin complex migrating at 95–100 kDa.
Concentrate fractions of interest at least 10-fold at 4 °C in a stirred-cell ultrafiltration unit fitted with an ultrafiltration disc membrane filter. Subsequently, subject the sample to buffer exchange by washing three times with 10 volumes of PBS containing 0.1% CHAPS in the same stirred cell system. Alternatively, concentrate fractions and exchange buffer using Amicon Ultracel 30 centrifugal filter columns at 4 °C according to the manufacturer’s instructions.
Determine protein concentration using a bicinchoninic acid (BCA) protein assay following the manufacturer's instructions. If required, samples can be further concentrated using an Amicon Ultracel 30 centrifugal filter at 4 °C according to the manufacturer's instructions.
Determine dentilisin proteolytic activity using the chromogenic substrate SAAPFNA as described previously [21].
Load 50 µL of each protein sample and controls in sample buffer into the wells of a 96-well plate (negative control: protein sample buffer; positive control: protein sample buffer plus chymotrypsin).
Incubate at 37 °C for 5 min while making up enzyme substrate.
Dilute 10× reaction buffer 1:5 in distilled H2O.
Dilute SAAPFNA substrate stock 1:50 in the resulting 2× reaction buffer.
Add 50 µL of SAAPFNA substrate to each well. Use sample buffer plus substrate as a blank.
Incubate plate at 37 °C for 5–30 min, monitoring carefully for yellow color change in positive wells vs. blank wells.
Read absorbance value at 405 nm.
Store purified dentilisin at -80 °C in aliquots appropriately sized for intended uses. In our hands, properly stored dentilisin samples are stable for several years.
Each step of dentilisin purification including growth, harvesting, detergent extraction/phase separation, and PC-PAGE is shown in Figure 1. Detection of protein products of the T. denticola prcB-prcA-prtP operon in whole cells and purified dentilisin protease complex are shown in Figure 2.
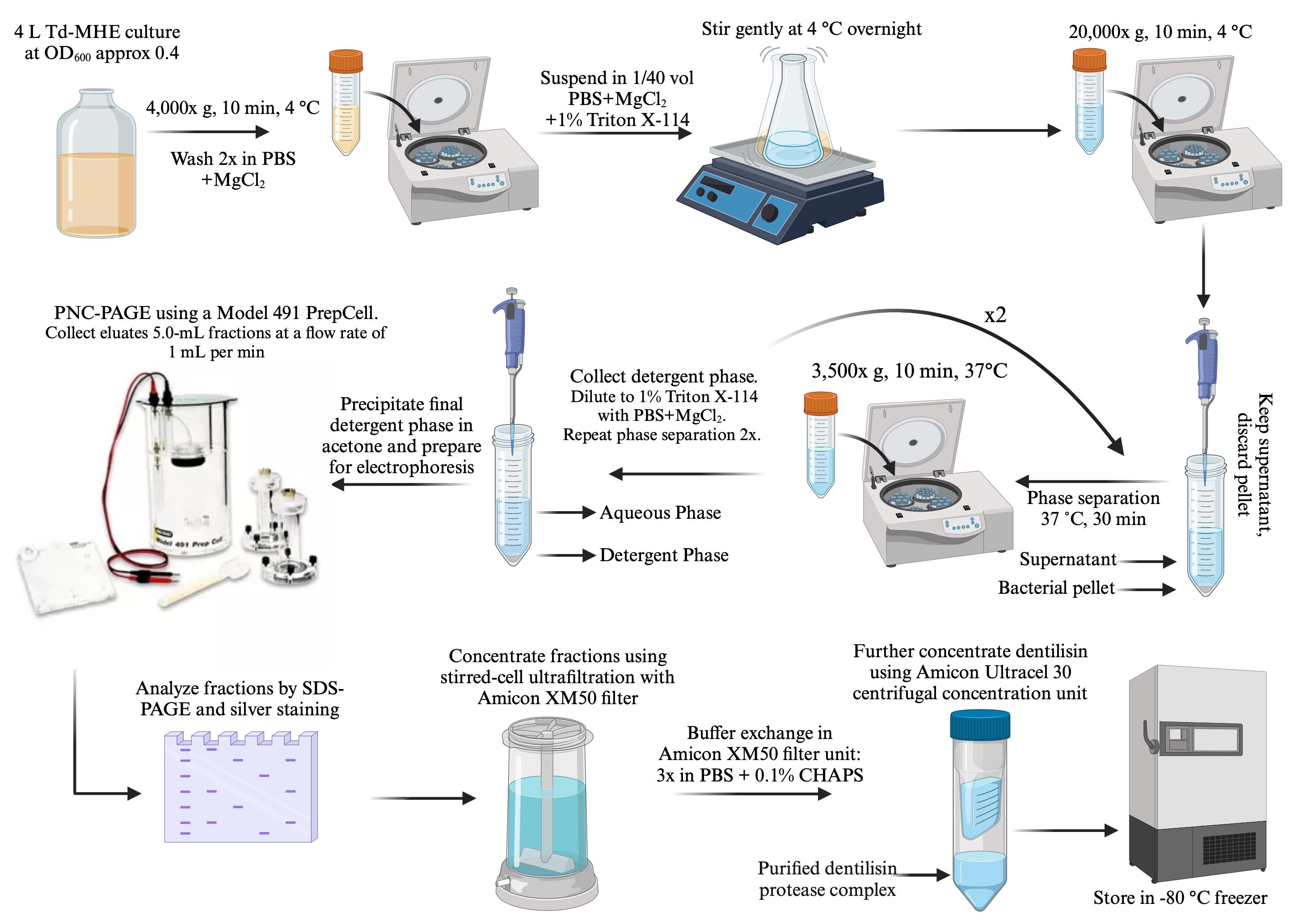
Figure 1. Dentilisin purification workflow. Details of each step, including growth, harvesting, detergent extraction/phase separation, and PC-PAGE, are described in the text.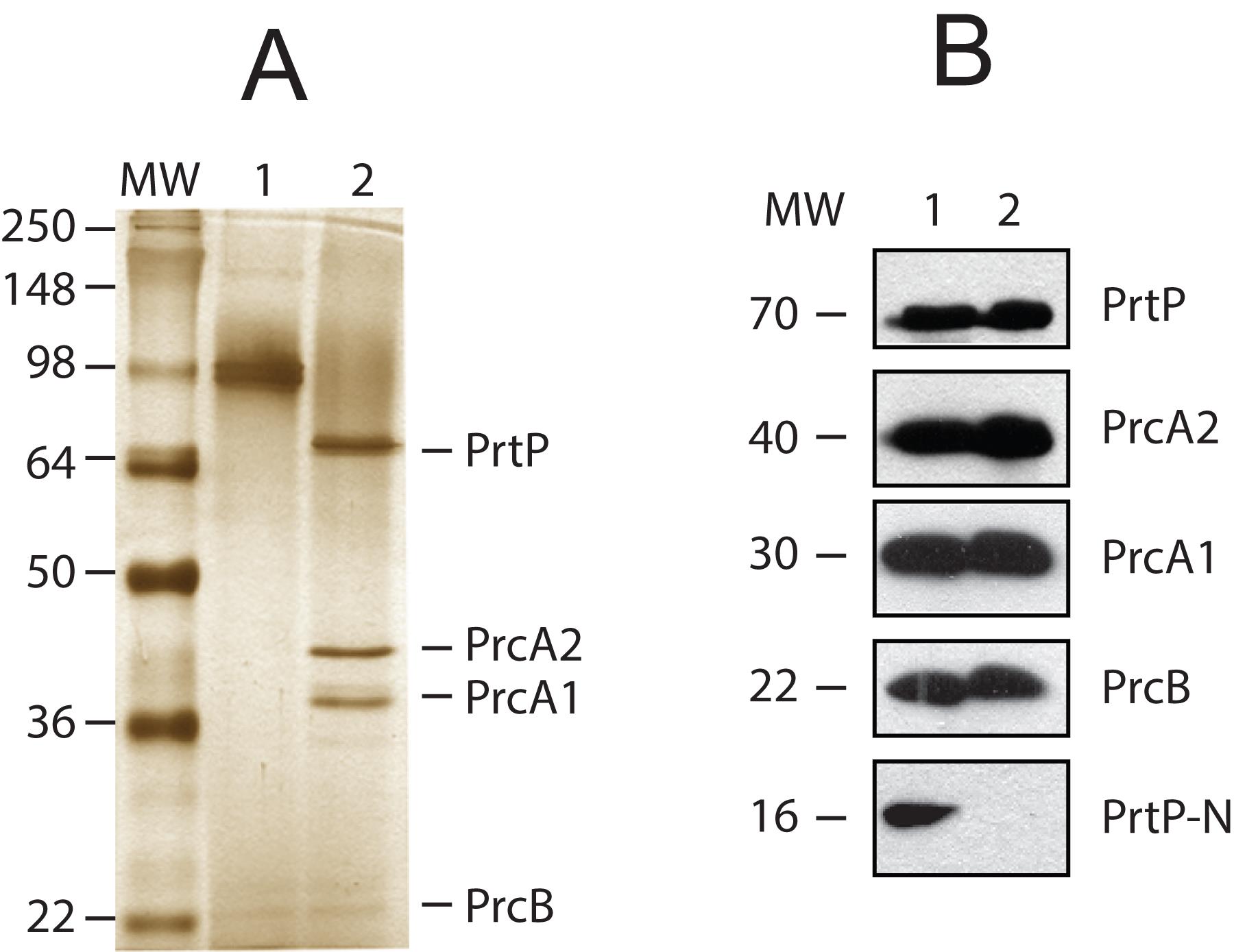
Figure 2. Detection of products of the Treponema denticola prcB-prcA-prtP operon in whole cells and purified dentilisin protease complex. A. Silver-stained SDS-PAGE of purified protease complex (0.4 µg per lane), unheated (lane 1) and boiled (lane 2). The locations of PrtP, PrcA1, PrcA2, and PrcB bands are indicated. Molecular weight standards in kDa (MW) are shown. B: Immunoblots of T. denticola whole-cell extracts and purified protease complex. All samples were boiled prior to electrophoresis. Samples were separated by standard SDS-PAGE and then transferred to nitrocellulose membranes. Lane 1: T. denticola whole-cell extract (equivalent to 10 µL of culture). Lane 2: purified dentilisin protease complex (0.4 µg per lane). Membranes were first probed with rabbit polyclonal antibodies specific for PrcB, PrcA1, PrcA2, PrtP-N-terminal domain, and mature PrtP [24,37,38] and then probed with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG. Protein bands of interest were visualized using the SuperSignal West Pico chemiluminescent substrate. The approximate size in kDa of each polypeptide is indicated (MW). Reprinted with permission from Godovikova et al. [24] (Figure 2) with minor modifications.
Part III: Gelatin zymography to detect activation of MMP-2 by T. denticola dentilisin
Native T. denticola dentilisin directly activates recombinant MMP-2
The oral spirochete Treponema denticola is highly associated with periodontal disease. The T. denticola outer membrane protein complex dentilisin contributes to the chronic activation of pro-MMP-2 in PDL cells and triggers increased expression levels of activators and effectors of active MMP-2 in PDL cells [14,39]. Despite these advances, there is no identified mechanism for dentilisin-induced MMP-2 activation. Gelatin zymography is an effective method for detecting gelatinolytic activity in various biological samples including culture media, cell extracts, tissue extracts, and biological fluids. This is accomplished by utilizing SDS-polyacrylamide gels that have been impregnated with gelatin. In this study, we explored the ability of T. denticola dentilisin to directly activate MMP-2. This is done by using both native purified T. denticola dentilisin and recombinant MMP2 in an in vitro setting, wherein MMP-2 activation is assayed by gelatin zymography and an in situ gelatin zymography assay.
A. Gelatin zymography
Materials and reagents
Reagents
Pierce BCA protein assay kit (Thermo Fisher, catalog number: 23225)
Pro-MMP-2 (R&D Systems, catalog number: 902-MP-010)
4× sample buffer (Bio-Rad, catalog number: 1610747)
10% Zymogram plus (gelatin) gel (Invitrogen, catalog number: ZY00100)
10× Gel electrophoresis running buffer (Novex, catalog number: LC2675)
Zymogram renaturing buffer (Novex, catalog number: LC2670)
Zymogram developing buffer (Novex, catalog number: LC2671)
Staining solution (Simplyblue safestain) (Invitrogen, catalog number: 465034)
Methanol (Fisher Scientific, catalog number: A452-4)
Acetic acid (Sigma-Aldrich, catalog number: A6283)
Solutions
Destaining solution (see Recipes)
Recipes
Destaining solution
10% methanol and 5% acetic acid in dH2O
Equipment
XCell SureLockTM electrophoresis cell (Thermo Fisher, model: EI0001)
PowerPacTM Basic (Bio-Rad, model: 041BR93879)
Eppendorf tubes (Eppendorf, catalog number: 0030 124.537)
ChemiDoc MP image system (Bio-Rad, model: Universal hood III, catalog number: 731BR01488)
Software and datasets
Image processing software (ImageJ version 2.14.0/1.54f, https://imagej.net/ij/download.html)
Procedure
Running and developing the gel
Conduct gelatin zymography as previously described [14].
Calculate protein concentrations for each sample using the Pierce BCA Protein Assay kit according to the manufacturer’s instructions.
Mix and incubate different concentrations of purified dentilisin and pro-MMP-2 for 30 min at RT and mix with 4× sample buffer. Then, load into each well and subject to SDS-PAGE on 10% gel containing gelatin.
After electrophoresis, remove SDS from the gels by washing them in renaturing buffer twice for 30 min. Then place gels in developing buffer for 30 min.
Replace the developing buffer and incubate gels at 37 for 16 h. Afterward, stain them with Simplyblue safestain for 30 min and subsequently destain using a solution of 5% acetic acid and 10% methanol until clear bands appear against a blue background.
Take zymogram pictures using ChemiDoc MP image system and measure the band intensity for active MMP-2 expression using ImageJ software (Figure 3).
For Figure 3, data was compared using one-way ANOVA Tukey’s multiple comparisons test. **p < 0.01; *** p < 0.001.
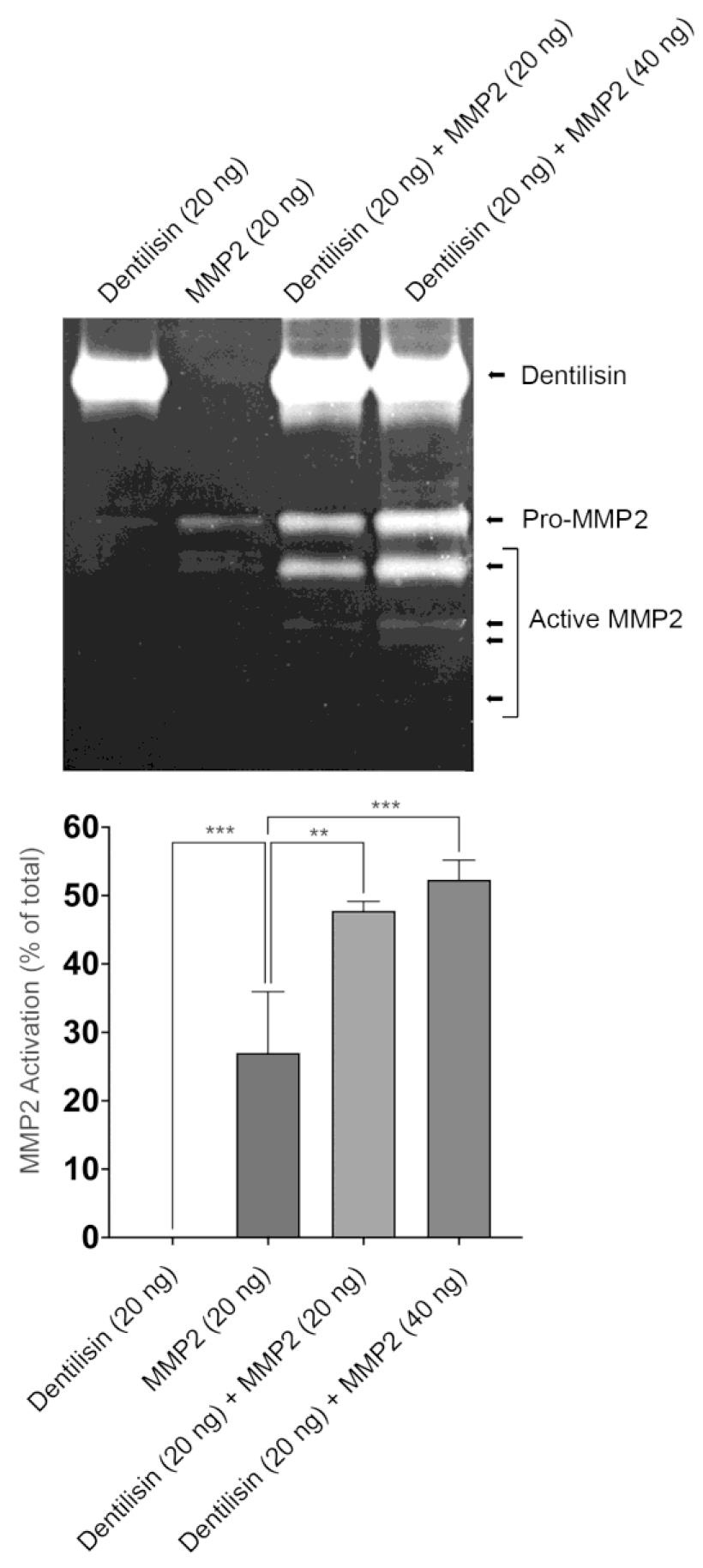
Figure 3. Dentilisin from Treponema denticola directly activates recombinant MMP-2. Top: Representative gelatin zymography image showing the activation of MMP-2. Bottom: The band intensity for active MMP-2 expression was measured using ImageJ software (n = 3). Data was compared using one-way ANOVA Tukey’s multiple comparisons test. **p < 0.01; ***= p < 0.001.
B. In situ gelatinase assay
Materials and reagents
Reagents
EnzChekTM gelatinase/collagenase assay kit (Invitrogen, catalog number: E12055)
Dye-quenched (DQ) gelatin fluorogenic substrate (Invitrogen, catalog number: E12054)
Pro-MMP-2 (R&D Systems, catalog number: 902-MP-010)
Pierce BCA protein assay kit (Thermo Fisher, catalog number: 23225)
Equipment
Fluorescent microplate reader (Molecular Devices, model: SPECTRA max, M2)
96-well plates (Thermo Fisher, catalog number: M33089)
Software and datasets
GraphPad Prism statistical software (GraphPad, Prism, version 10.1.1)
Procedure
Add 80 mL of 1× reaction buffer and 20 mL of the 1 mg/mL DQ gelatin fluorogenic substrate stock to each assay well.
Test different sample groups with control, purified pro-MMP-2, purified dentilisin, or both pro-MMP-2 and dentilisin with gelatin substrate and reaction buffer. Use Pierce’s BCA protein quantification kit according to the manufacturer’s instructions to determine the initial concentration of 1 mg/mL for each purified protein.
Achieve further dilution to a final concentration of 0.002 mg/mL for each protein with reaction buffer similarly to a previously described publication [40].
Add 100 mL of the final concentration of 1× reaction buffer (negative control), MMP-2 alone, or dentilisin alone to respective grouped wells.
Add 50 mL of the final concentration MMP-2 plus 50 mL of final concentration dentilisin to its respective well group.
Read samples in a fluorescent microplate reader at 495 nm absorption and 515 nm emission at 0, 30, 60, 90, and 120 min time points.
Data represents mean ± SD from two independent experiments (Figure 4).
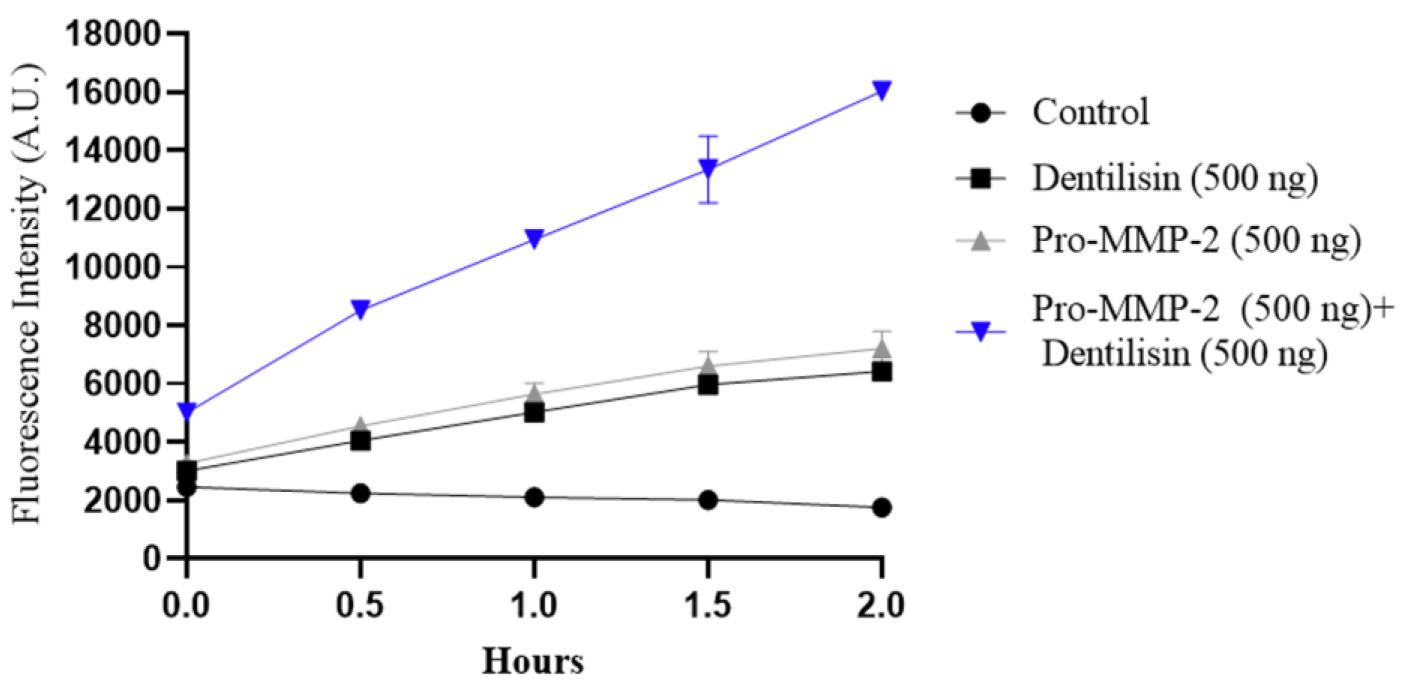
Figure 4. Direct activation of pro-MMP-2 by dentilisin using in situ gelatin zymography. Fluorescent intensity of each group was plotted at each interval (0, 30, 60, 90, and 120 min). Sample groups were negative control, purified pro-MMP-2 alone, purified dentilisin, or both pro-MMP-2 and dentilisin. The plot represents an average of two experiments, each consisting of six separate samples in every group.
Data analysis
Data was compared using one-way ANOVA with Tukey’s multiple comparisons test (Figure 3, Statistical analysis, Front Cell Infect Microbiol. 2021 May 19; 11:671968).
Validation of protocol
The protocols for dentilisin purification and zymography described here have been previously reported and validated in:
Fenno et al. [20]. Infect. Immun. 66:1869–1877. doi: 10.1128/IAI.66.5.1869-1877.1998 (Figure 3).
Godovikova et al. [24]. Infect. Immun. 79(12):4868–75. doi: 10.1128/IAI.05701-11 (Figure 2).
Miao et al. [14]. Infect. Immun. 79(2):806–11. doi: 10.1128/IAI.01001-10 (Figure 2, 3B, 4A, 5A).
Miao et al. [39]. Arch. Oral Biol. 59(10):1056–64. doi: 10.1016/j.archoralbio.2014.06.003 (Figure 1A and B, 3C).
Malone et al. [17]. Front. Cell Infect. Microbiol. 11:671968. doi: 10.3389/fcimb.2021.671968 (Figure 4, 7A, 8A).
Ganther et al. [16]. PLoS Pathog. 17(7): doi: 10.1371/journal.ppat.1009311 (Figure 4b, S2).
Acknowledgments
These studies were supported by funding from the NIH as follows: R01 DE025225 to YLK and JCF; R01 DE018221 to JCF; Ruth L. Kirschstein Individual Predoctoral NRSA for MD/PhD, and other Dual Degree Fellowships (F30 DE027598) to EM; and Ruth L. Kirschstein NRSA Institutional Research Training Grant (T32DE007306) to SG. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of this manuscript. The protocols described here were adapted from previously reported studies by the authors [14–16,20 22,24].
Competing interests
The authors declare no conflict of interest.
References
- Cunningham, T. M., Walker, E. M., Miller, J. N. and Lovett, M. A. (1988). Selective release of the Treponema pallidum outer membrane and associated polypeptides with Triton X-114. J. Bacteriol. 170(12): 5789–5796.
- Socransky, S., Haffajee, A., Cugini, M., Smith, C. and Kent, R. L. (1998). Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25(2): 134–144.
- Marsh, P. D. (2003). Are dental diseases examples of ecological catastrophes? Microbiology 149(2): 279–294.
- Darveau, R. P. (2010). Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8(7): 481–490.
- Schulz, S., Porsch, M., Grosse, I., Hoffmann, K., Schaller, H. G. and Reichert, S. (2019). Comparison of the oral microbiome of patients with generalized aggressive periodontitis and periodontitis-free subjects. Arch. Oral Biol. 99: 169–176.
- Ai, D., Huang, R., Wen, J., Li, C., Zhu, J. and Xia, L. C. (2017). Integrated metagenomic data analysis demonstrates that a loss of diversity in oral microbiota is associated with periodontitis. BMC Genomics 18(Suppl 1): 1041.
- Choi, B. K., Paster, B. J., Dewhirst, F. E. and Göbel, U. B. (1994). Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 62(5): 1889–1895.
- Ellen, R. P. and Galimanas, V. B. (2005). Spirochetes at the forefront of periodontal infections. Periodontology 2000 38(1): 13–32.
- Simonson, L. G., Goodman, C. H., Bial, J. J. and Morton, H. E. (1988). Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect. Immun. 56(4): 726–728.
- Yoshida, A., Kawada, M., Suzuki, N., Nakano, Y., Oho, T., Saito, T. and Yamashita, Y. (2004). TaqMan real‐time polymerase chain reaction assay for the correlation of Treponema denticola numbers with the severity of periodontal disease. Oral Microbiol. Immunol. 19(3): 196–200.
- Goetting-Minesky, M. P., Godovikova, V. and Fenno, J. C. (2021). Approaches to Understanding Mechanisms of Dentilisin Protease Complex Expression in Treponema denticola. Front. Cell. Infect. Microbiol. 11: e668287.
- Chi, B., Qi, M. and Kuramitsu, H. K. (2003). Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res. Microbiol. 154(9): 637–643.
- Uitto, V. J., Pan, Y. M., Leung, W. K., Larjava, H., Ellen, R. P., Finlay, B. B. and McBride, B. C. (1995). Cytopathic effects of Treponema denticola chymotrypsin-like proteinase on migrating and stratified epithelial cells. Infect. Immun. 63(9): 3401–3410.
- Miao, D., Fenno, J. C., Timm, J. C., Joo, N. E. and Kapila, Y. L. (2011). The Treponema denticola Chymotrypsin-Like Protease Dentilisin Induces Matrix Metalloproteinase-2-Dependent Fibronectin Fragmentation in Periodontal Ligament Cells. Infect. Immun. 79(2): 806–811.
- Ateia, I. M., Sutthiboonyapan, P., Kamarajan, P., Jin, T., Godovikova, V., Kapila, Y. L. and Fenno, J. C. (2018). Treponema denticola increases MMP-2 expression and activation in the periodontium via reversible DNA and histone modifications. Cell. Microbiol. 20(4): e12815.
- Ganther, S., Radaic, A., Malone, E., Kamarajan, P., Chang, N. Y., Tafolla, C., Zhan, L., Fenno, J. C. and Kapila, Y. L. (2021). Treponema denticola dentilisin triggered TLR2/MyD88 activation upregulates a tissue destructive program involving MMPs via Sp1 in human oral cells. PLoS Pathog. 17(7): e1009311.
- Malone, E. T., Ganther, S., Mena, N., Radaic, A., Shariati, K., Kindberg, A., Tafolla, C., Kamarajan, P., Fenno, J. C., Zhan, L., et al. (2021). Treponema denticola-Induced RASA4 Upregulation Mediates Cytoskeletal Dysfunction and MMP-2 Activity in Periodontal Fibroblasts. Front. Cell. Infect. Microbiol. 11: e671968.
- McDowell, J., Frederick, J., Miller, D., Goetting‐Minesky, M., Goodman, H., Fenno, J. and Marconi, R. (2010). Identification of the primary mechanism of complement evasion by the periodontal pathogen, Treponema denticola. Mol. Oral Microbiol. 26(2): 140–149.
- Ellen, R. P., Song, M. and McCulloch, C. A. (1994). Degradation of endogenous plasma membrane fibronectin concomitant with Treponema denticola 35405 adhesion to gingival fibroblasts. Infect. Immun. 62(7): 3033–3037.
- Fenno, J. C., Hannam, P. M., Leung, W. K., Tamura, M., Uitto, V. J. and McBride, B. C. (1998). Cytopathic Effects of the Major Surface Protein and the Chymotrypsinlike Protease of Treponema denticola. Infect. Immun. 66(5): 1869–1877.
- Grenier, D., Uitto, V. J. and McBride, B. C. (1990). Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect. Immun. 58(2): 347–351.
- Fenno, J. C., Tamura, M., Hannam, P. M., Wong, G. W. K., Chan, R. A. and McBride, B. C. (2000). Identification of a Treponema denticola OppA Homologue That Binds Host Proteins Present in the Subgingival Environment. Infect. Immun. 68(4): 1884–1892.
- Godovikova, V., Goetting-Minesky, M. P., Timm, J. C. and Fenno, J. C. (2019). Immunotopological Analysis of the Treponema denticola Major Surface Protein (Msp). J. Bacteriol. 201(2): e00528–18.
- Godovikova, V., Goetting-Minesky, M. P. and Fenno, J. C. (2011). Composition and Localization of Treponema denticola Outer Membrane Complexes. Infect. Immun. 79(12): 4868–4875.
- Fenno, J. (1998). Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponemadenticola. FEMS Microbiol. Lett. 163(2): 209–215.
- Fenno, J. C. (2006). Laboratory Maintenance of Treponema denticola. Curr. Protoc. Microbiol. Chapter 12:Unit 12B.1. doi: 10.1002/9780471729259.mc12b01s00.
- Ohta, K., Makinen, K. K. and Loesche, W. J. (1986). Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect. Immun. 53(1): 213–220.
- Bordier, C. (1981). Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256(4): 1604–1607.
- Rosenthal, K. S. and Koussaie, F. (1983). Critical micelle concentration determination of nonionic detergents with Coomassie Brilliant Blue G-250. Anal. Chem. 55(7): 1115–1117.
- Haake, D. A., Martinich, C., Summers, T. A., Shang, E. S., Pruetz, J. D., McCoy, A. M., Mazel, M. K. and Bolin, C. A. (1998). Characterization of Leptospiral Outer Membrane Lipoprotein LipL36: Downregulation Associated with Late-Log-Phase Growth and Mammalian Infection. Infect. Immun. 66(4): 1579–1587.
- Sela, M. N., Bolotin, A., Naor, R., Weinberg, A. and Rosen, G. (1997). Lipoproteins of Treponema denticola: their effect on human polymorphonuclear neutrophils. J. Periodontal Res. 32(5): 455–466.
- Carroll, J. A. (2010). Methods of Identifying Membrane Proteins in Spirochetes. Curr. Protoc. Microbiol. Chapter 12:Unit12C 2. doi: 10.1002/9780471729259.mc12c02s16.
- Crother, T. R. and Nally, J. E. (2008). Analysis of Bacterial Membrane Proteins Produced During Mammalian Infection Using Hydrophobic Antigen Tissue Triton Extraction (HATTREX). Curr. Protoc. Microbiol. Chapter 12:Unit 12 1. doi: 10.1002/9780471729259.mc1201s9.
- Holt, S. C. and Canale-Parola, E. (1968). Fine Structure of Spirochaeta stenostrepta, a Free-living, Anaerobic Spirochete. J. Bacteriol. 96(3): 822–835.
- Uitto, V. J., Grenier, D., Chan, E. C. and McBride, B. C. (1988). Isolation of a chymotrypsinlike enzyme from Treponema denticola. Infect. Immun. 56(10): 2717–2722.
- Ishihara, K., Miura, T., Kuramitsu, H. K. and Okuda, K. (1996). Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin). Infect. Immun. 64(12): 5178–5186.
- Godovikova, V., Wang, H. T., Goetting-Minesky, M. P., Ning, Y., Capone, R. F., Slater, C. K. and Fenno, J. C. (2010). Treponema denticola PrcB Is Required for Expression and Activity of the PrcA-PrtP (Dentilisin) Complex. J. Bacteriol. 192(13): 3337–3344.
- Lee, S. Y., Bian, X. L., Wong, G. W. K., Hannam, P. M., McBride, B. C. and Fenno, J. C. (2002). Cleavage of Treponema denticola PrcA Polypeptide To Yield Protease Complex-Associated Proteins Prca1 and Prca2 Is Dependent on PrtP. J. Bacteriol. 184(14): 3864–3870.
- Miao, D., Godovikova, V., Qian, X., Seshadrinathan, S., Kapila, Y. L. and Fenno, J. C. (2014). Treponema denticola upregulates MMP-2 activation in periodontal ligament cells: Interplay between epigenetics and periodontal infection. Arch. Oral Biol. 59(10): 1056–1064.
- Sarker, H., Hardy, E., Haimour, A., Maksymowych, W. P., Botto, L. D. and Fernandez-Patron, C. (2019). Identification of fibrinogen as a natural inhibitor of MMP-2. Sci. Rep. 9(1): 4340.
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Kamarajan, P., Timm, J. C., Goetting-Minesky, M. P., Malone, E. T., Ganther, S., Radaic, A., Tafolla, C., Fenno, J. C. and Kapila, Y. L. (2024). Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography. Bio-protocol 14(7): e4970. DOI: 10.21769/BioProtoc.4970.
Category
Microbiology > Microbe-host interactions > Bacterium
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









