- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Transient Expression Assay and Microscopic Observation in Kumquat Fruit
Published: Vol 14, Iss 7, Apr 5, 2024 DOI: 10.21769/BioProtoc.4968 Views: 1756
Reviewed by: Wenrong HeXiongjie ZhengMin Cao

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry
Sakharam Waghmare [...] Rucha Karnik
Nov 20, 2025 2025 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views
Abstract
Citrus fruits encompass a diverse family, including oranges, mandarins, grapefruits, limes, kumquats, lemons, and others. In citrus, Agrobacterium tumefaciens–mediated genetic transformation of Hongkong kumquat (Fortunella hindsii Swingle) has been widely employed for gene function analysis. However, the perennial nature of woody plants results in the generation of transgenic fruits taking several years. Here, we show the procedures of Agrobacterium-mediated transient transformation and live-cell imaging in kumquat (F. crassifolia Swingle) fruit, using the actin filament marker GFP-Lifeact as an example. Fluorescence detection, western blot analysis, and live-cell imaging with confocal microscopy demonstrate the high transformation efficiency and an extended expression window of this system. Overall, Agrobacterium-mediated transient transformation of kumquat fruits provides a rapid and effective method for studying gene function in fruit, enabling the effective observation of diverse cellular processes in fruit biology.
Graphical overview
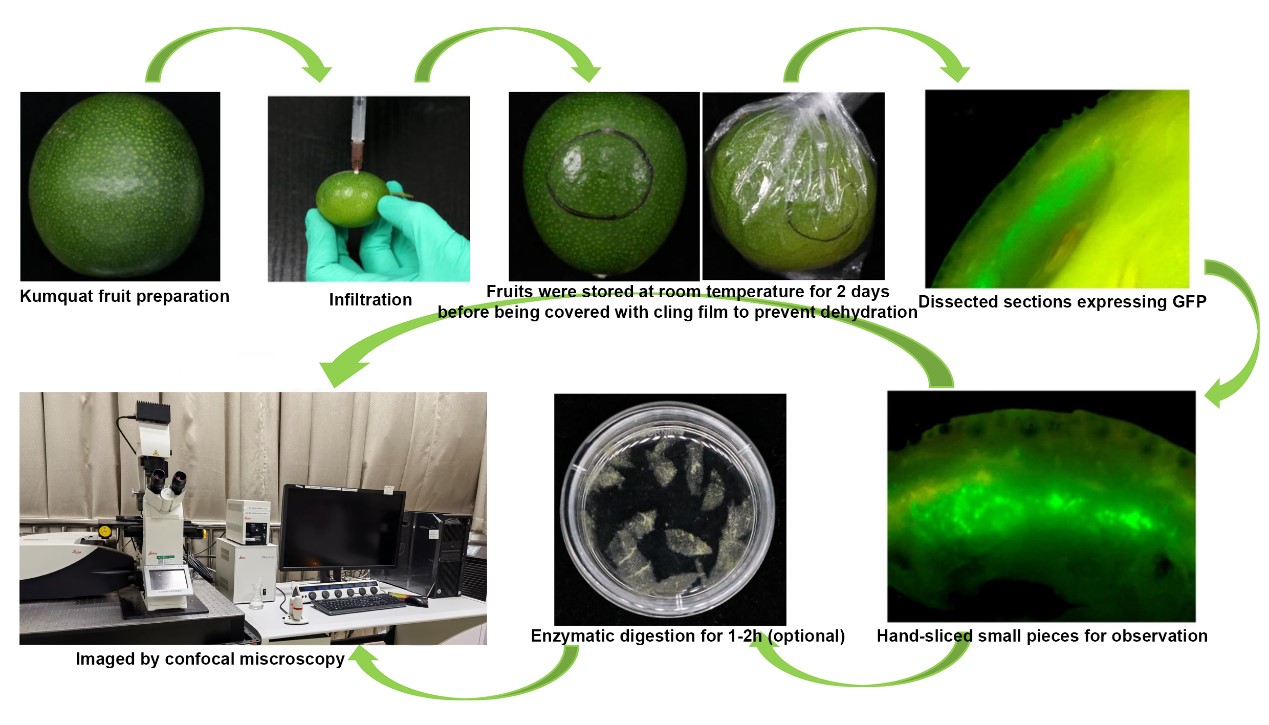
Background
Citrus, one of the most important fruit crops in the world, is subjected to a variety of environmental stimuli and developmental signals during its growth. Observing the dynamics of organelles may provide new insights into the vital behavior of fruits in response to these developmental and environmental signals. The use of green fluorescent protein (GFP) and its derivatives is a great improvement for cell biological studies, as it can be fused to genes of interest to study their subcellular localization, dynamics, and protein–protein interactions [1]. However, cell structures and subcellular activities from citrus fruits can hardly be observed by light microscopy due to their 3D structure and little transparency. The lack of efficient transformation techniques and the long juvenile phase of citrus plants [2] make the study of cell biology even more difficult. Therefore, developing an effective method for transient transformation of citrus species is important. Agrobacterium-mediated infiltration of tobacco leaves was an early-developed method for transient expression [3] and has been widely used to study cell morphology and dynamics. Subsequently, Agrobacterium-mediated transient expression methods have been developed for a wide range of leaves and fruits including Arabidopsis [4], tomato [5], strawberry [6], apple [7,8], and also citrus fruits [9,10]. Here, the kumquat (Fortunella crassifolia Swingle) fruit has been selected for Agrobacterium-mediated transient expression. It is a major citrus cultivar in southern China, bearing smaller fruits in the Citrus genus compared to pomelo, oranges, and tangerines [11]. It is also characterized by its high sugar content, thin skin, and fewer juice cells, which provides a good environment for infiltration and infection by Agrobacterium [12].
In eukaryotic cells, the cytoskeleton is a network of protein–fiber reticulum consisting of microtubules, microfilaments, and intermediate filaments, which not only plays an important role in maintaining cell morphology, withstanding external forces, and maintaining the orderliness of the internal cell structure, but also participates in many important biological processes. Hence, imaging this network in its native and mobile states is of great importance. Here, using the GFP-Lifeact for actin cytoskeleton as an example, we describe methods for Agrobacterium--mediated transient transformation of citrus fruit and live-cell imaging in citrus fruit. The high expression of fluorescent protein in kumquat fruit, combined with subsequent live cell imaging, provides a method for studying gene function, subcellular localization, and cellular activity in fruit.
Materials and reagents
Biological materials
Kumquat (F. crassifolia Swingle) fruit (collected from Guangxi Institute of Citrus Research, located at Guilin city, Guangxi, China)
Reagents
Actin cytoskeleton marker GFP-Lifeact containing a 35S::GFP cassette as a visual reporter [13]
P19 Agrobacterium
Yeast extract (OXOID, catalog number: LP0021)
Tryptone (OXOID, catalog number: LP0042)
NaCl (HUSHI, catalog number: 10019318, CAS: 7647-14-5)
D-glucose (HUSHI, catalog number: 10010518, CAS: 14431-43-7)
MES (Sigma-Aldrich, catalog number: M8250)
Na3PO4 (HUSHI, catalog number: 20040928, CAS: 7601-54-9)
Acetosyringone (Sigma-Aldrich, catalog number: D134406)
GV3101 Chemically Competent Cell (Shanghai Weidi Biotechnology, catalog number: AC1001)
Antibiotics: Kanamycin (Kan) (Genview, catalog number: AK177-10G) and Rifampicin (Rif) (Genview, catalog number: AR280-1G)
Antibodies: Polyclonal mouse antibody against GFP (Biorbyt, catalog number: orb688437) as the primary antibody and goat anti-mouse HRP (Biorbyt, catalog number: orb670233) as the secondary antibody
Dimethyl sulfoxide (DMSO) (MACKLIN, catalog number: C13178602)
Mannitol (Solarbio, catalog number: M8141)
KCl (Rhawn, catalog number: R051965)
CaCl2 (MACKLIN, catalog number: C832203)
BSA (MACKLIN, catalog number: B928042)
Cellulase R10 (Yakult, catalog number: 220908-02)
Sodium hypochlorite (MACKLIN, catalog number: S935360)
Solutions
Luria Broth (LB) medium (see Recipes)
Infiltration buffer (see Recipes)
Enzymatic solution (see Recipes)
Antibiotics (see Recipes)
Recipes
LB media
10 g of tryptone, 10 g of NaCl, 5 g of yeast extract; make up to 1 L with dH 2O, following autoclave sterilization.
Infiltration buffer
Infiltration medium stock solutions:
500 mM MES: 4.88 g of MES in 50 mL of dH2O. Store at 4 °C.
20 mM Na3PO4: 0.17 g of Na3PO4 in 50 mL of dH2O. Store at 4 °C.
1 M Acetosyringone (3’,5’-dimethoxy-4’-hydroxyacetophenone): 0.196 g of Acetosyringone in 1 mL of DMSO. Divide into single-use aliquots and store at -20 °C.
50 mL infiltration buffer
250 mg of D-glucose, 5 mL of MES stock solution, 5 mL of Na3PO 4 stock solution, 5 μL of Acetosyringone stock solution; make up to 50 mL with dH2O.
Enzymatic solution
Enzymatic solution stock solutions:
0.8 M mannitol stock: 7.29 g of mannitol in 50 mL of dH2O.
2 M KCl stock: 7.455 g of KCl in 50 mL of dH2O.
0.2 M MES stock: 1.952 g of MES in 50 mL of dH2O. Store at 4 °C.
1 M CaCl2 stock: 1.11 g of CaCl2 in 10 mL of dH2 O.
10% BSA stock: 0.1 g of BSA in 1 mL of dH2O.
10 mL enzymatic solution: 0.15 g of cellulase R10, 0.04 g of macerozyme R1, 5 mL of 0.8 M mannitol stock, 100 μL of 2 M KCl stock, and 1 mL of 0.2 M MES (pH 5.7) stock. Heat solution in heat block or oven set to 55 °C (maximum) for 10 min to help enzymes dissolve (or place in 55 °C water) and then cool to room temperature (RT). Next, add 100 μL of 1 M CaCl 2 stock, 100 μL of 10% BSA stock, and dH2O up to 10 mL. Sterilize by filtration (0.45 μm).
Antibiotics
50 mg/L Rif
50 mg/L Kan
Laboratory supplies
Tips (Axygen, catalog number: AXYT1000B)
1 mL syringe with needle (GEMTIER, catalog number: 0.45X16 RW LB)
Glass microscope slides, coverslips (Corning, catalog number: CLS294875X25, CLS285522), and Vectashield (Vectorlabs, catalog number: H-1000) for microscopy
Test tubes (Eppendorf, catalog number: EP0030122178) and flask (Corning, catalog number: CLS4444250) for culturing Agrobacterium
Fine forceps and razor blades (Artis Tweezer, catalog number: Z742671; smartSlicer, catalog number: Z740503)
Marker pen (STATMARK, catalog number: Z648191)
Tray (PureSolv, catalog number: Z681687) and Falcon tube cap (Corning, catalog number: 352070) for placing fruit
Petri dishes (60 mm, optional, for enzymatic digestion of protoplasts) (Nalgene, catalog number: TMO5921-0060)
Pipette (Eppendorf, catalog number: GN686271)
Rubber gloves (Microflex, catalog number: Z265179)
Bibulous paper (for uptaking the extravasated Agrobacterium)
50 mL Falcon tubes (Corning, catalog number: 352070)
Equipment
Shaking incubator (Radobio, model: Stab M1T) used for the growth of Agrobacterium at 28 °C
Centrifuge (Eppendorf, model: 5424)
Clean workbench (AIRTECH, model: SW-CJ-2FD)
Fluorescence dissecting stereomicroscope (Olympus, model: SZX7)
Confocal microscope (Leica, model: SP8)
Nanodrop spectrophotometer (or equivalent) to determine optical density of bacterial culture
Procedure
Preparation of kumquat fruit for infestation
Carry out transformation of citrus fruit on the kumquat (see Note 1). Harvest “Huapi” kumquats or “Rongan” kumquats 150–210 days after flowering (see Note 2). Sterilize with 2% sodium hypochlorite solution for 2 min and rinse thoroughly in water; then, drain on bibulous paper before Agrobacterium injection.
Choose the healthy and fresh fruit for infiltration.
Agrobacterium preparation
Incubate Agrobacterium strain GV3101 (see Note 3) carrying GFP-Lifeact and P19 in liquid LB medium with Kan and Rif antibiotics overnight at 28 °C with shaking at 220 rpm for 12–16 h, to an optical density (O.D.) over 1.0.
Centrifuge Agrobacterium culture in a 50 mL Falcon tube at 5,000× g for 5 min to pellet the cells.
Discard the supernatant and resuspend Agrobacterium pellet using infiltration buffer.
Spin down the culture at 5,000× g for 5 min and discard the supernatant. Suspend the culture with the infiltration buffer to reach a final OD600.
Measure the OD600 of Agrobacterium cells and add appropriate volumes of infiltration buffer to dilute the Agrobacteria to the desired O.D. (usually 0.8 O.D., see Note 4). If you want to co-express two or multiple proteins, mix different kinds of Agrobacteria with the desired O.D. for each one. Every construct needs to be mixed in equal amounts with P19 Agrobacterium (see Note 5).
Agrobacterium infiltration
Cut off part of the needle of the syringe, leaving a needle approximately 0.1–0.3 cm in length.
Hold the bottom of the fruit in one hand and gently inject the bacteria solution into the epicarp at a depth of 0.1–0.3 cm. With gentle pressure on the plunger, carefully inject bacteria mixture into the fruit tissues (Video 1). Try to be as gentle as possible when injecting to avoid damaging the fruit. Inject each fruit with 0.2–0.5 mL of infiltration solution (see Note 6). The mixture could potentially distribute to an area with a radius of 0.8–1.3 cm from the injection point (data not shown). The infiltrated area can easily be seen as a dark, water-soaked region.
Get rid of the overflowing Agrobacterium on the surface of the fruit with bibulous paper. Mark the injection site with a marker pen to facilitate sampling (Video 1).
Video 1. Procedure for fruit injectionPlace the infiltrated fruit on the cap of 15/50 mL Falcon tubes and store at RT (25 °C) for approximately two days. Then, wrap with cling film to prevent dehydration (see Note 7) and continue to store at RT.
Observation of fluorescent signals
Approximately five days after infiltration (see Note 8), swiftly dissect the tissue infiltrated with Agrobacterium using a sharp blade (underscored area), with no particular concern for the size or thickness of tissue blocks. Subsequently, place these tissue blocks gently on a microscopy slide and immediately observe under a stereomicroscope equipped with a DP70 camera. Fluorescent signals near the injection site are distinctly visible (see Note 9). The purpose of this step is to preliminarily assess the expression, intensity, and tissue localization of the target protein through fluorescent signals, facilitating subsequent sampling. Detailed subcellular imaging, providing information on cellular localization, necessitates confocal microscopy.
Confocal microscopy
Hand-slice tissues with strong fluorescence signals into small pieces, as thin as possible, and transfer to a slide for confocal microscopy directly (see Note 10).
(Optional if you need to use protoplasts for microscopic observation) Incubate samples for confocal microscopy in sterilized enzymatic solution for 1–2 h in the dark (see Note 11) with shaking at 40 rpm at room temperature to partially digest the cell wall. The main purpose of this additional step is to release the cells from the tissue to facilitate observation. Prolonged digestion can result in the production of protoplasts, but it is normally unnecessary; ideally, it is good to keep any disruptions of this process to a minimum (Observation of Golgi apparatus movement using protoplasts is shown in Video 2).
Video 2. Golgi movement in citrus fruit protoplastMount samples in microscope slides and seal with Vectashield; then, press the coverslip gently from one side.
Carry out live-cell imaging using a Leica SP8 laser scanning confocal microscope with a 63× oil immersion lens. For wavelength settings, GFP was excited at 488 nm and detected at 505–550 nm.
Data analysis
“Huapi” or “Rongan” kumquat fruits (Figure 1A) were used for the transient overexpression of GFP-Lifeact. Five days after the injection, the injection site was incised, and obvious green fluorescence was visible under a stereomicroscope (Figure 1B). Fluorescent signals could be detected in different tissues of the kumquat, including the flavedo, albedo, juice sacs, and partition, into which the Agrobacterium solution penetrated [12]. Western blot analysis conducted on kumquat tissues five days post-injection revealed that the expression of GFP-Lifeact was discernible at all Agrobacterium OD values exceeding 0.2. Furthermore, the protein expression level of GFP-Lifeact exhibited enhancement with escalating concentrations of Agrobacterium injection (Figure 1C). Tissues exhibiting robust fluorescent signals were meticulously sectioned as thinly as possible, and the GFP-Lifeact-labeled actin cytoskeletons were clearly observed under confocal microscopy through manual sections and protoplasts (Figure 1D). However, the free GFP is ubiquitously distributed throughout the fruit cells, with signals of free GFP detectable in both the membrane and cytoplasm (Figure 2).
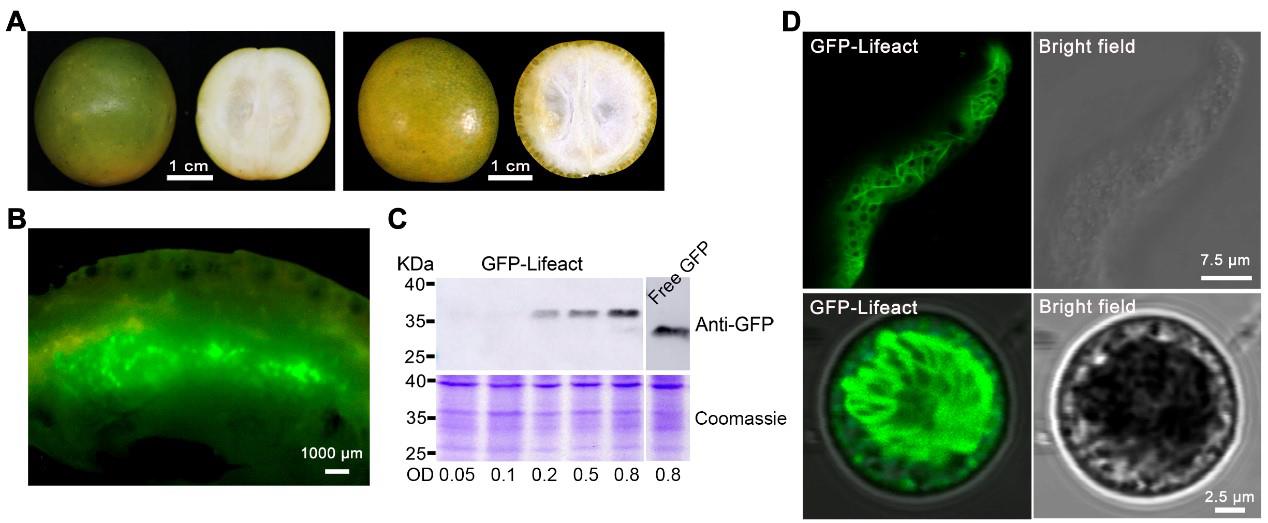
Figure 1. Transient expression of fluorescent protein GFP-Lifeact in kumquat fruit cells. A. Representative image of “Huapi” (left) and “Rongan” (right) kumquat fruit used for injection. B. Dissected sections expressing GFP fusion proteins. C. Western blotting analysis of GFP-Lifeact and free GFP (from pMDC43 binary vector) expression in kumquat fruit using different concentrations of Agrobacterium suspension; 20 μg of total protein from each lane were probed with anti-GFP antibodies with 1:2,000 dilution. D. Representative images of fluorescent protein fusions localized to actin cytoskeleton in fruit cells through manual sectioning (up) and protoplasts (down). The construct GFP-Lifeact was infiltrated at OD600 = 0.8 and images were taken five days after infiltration.
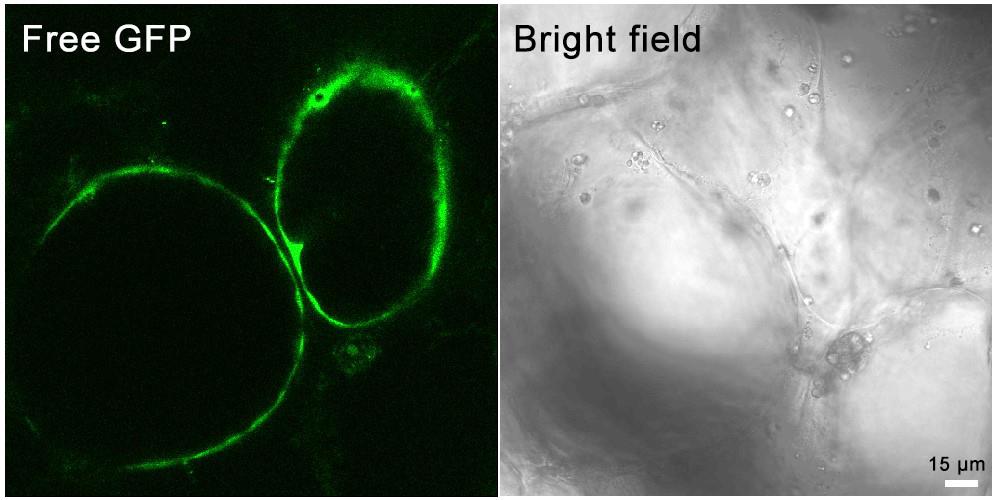
Figure 2. Transient expression of the free GFP in kumquat fruit cells
Notes
Fruit variety selection: The meticulous choice of fruit varieties holds paramount significance. Kumquat fruits were identified as our preferred material for transient transformation due to their characteristics of thin skin and tender flesh (Figure 1A); additionally, they contain high sugar content and low acid, which provides a suitable growth environment for Agrobacterium. Other citrus cultivars, such as orange, pomelo, and mandarin, could also be considered. Notably, kumquat exhibited sustained production of robust signals and high conversion efficiency (data not shown).
Determination of the fruit stage: The mature green stage of kumquat fruit (approximately 150–210 days after flowering) has proven to be optimal for transient transformation, owing to its firm texture, heightened metabolic activity, and finer peels. Additionally, post-harvest kumquat fruit remains suitable for experimentation, underscoring the importance of selecting fresh citrus fruits for injection in subsequent procedures. It is advised to avoid fruit at the end of the ripening stage, as it is susceptible to rot following Agrobacterium injection.
Strain selection: Various strains of Agrobacterium tumefaciens (e.g., EHA105 or GV3101) were selected for testing. The findings revealed that transformation mediated by GV3101 yielded the highest protein output.
Optical density value: Attaining the optimal Agrobacterium density (OD600) for each construct is crucial, and this requirement varies significantly from gene to gene. The OD600 of Agrobacterium also exerts an impact on the level of protein expression, necessitating the optimization of the ideal OD600 for a new construct prior to the commencement of the actual study. An OD600 concentration of Agrobacterium below 0.1 typically results in weak expression, while an OD600 exceeding 1.0 tends to induce yellowing and rotting of fruit. If mitigating potential overexpression artifacts is a primary concern, it is advisable to use the lowest O.D. that still yields sufficient signal. Otherwise, an OD600 within the range of 0.5–0.8 can be employed to achieve maximum expression.
P19: Augmented expression can be attained through the utilization of p19 constructs, effectively averting gene silencing.
Injection dose: Maintaining the injection volume within the range of 0.2–0.5 mL for each fruit is imperative, as excessive infiltration buffer may lead to unforeseen rotting.
Bagging: After two days of fruit injection, it is advisable to individually encase fruits with cling film to enhance water retention and ensure air permeability; bagging should not occur too early, otherwise it will cause fruit rot due to high humidity.
Time of expression: Different proteins may exhibit distinct expression timelines. Generally, protein expression becomes detectable 4–5 days after infiltration. If the fruit preservation conditions are favorable, protein expression can even be detected up to one month post-infiltration. The persistence and abundance of protein expression typically depend on the plasmid itself and the vitality of the fruits.
Fruit tissues for expression: Fluorescent signals can be observed in various tissues of kumquat, encompassing flavedo, albedo, and juice sacs, where the Agrobacterium solution can penetrate. The efficacy of expression is contingent upon the uptake of Agrobacterium by different tissues, with superior absorption correlating to enhanced expression strength. Typically, the expression of fluorescent proteins in albedo surpasses that in other tissues.
During the process of slicing fruit tissue, it is imperative to minimize external pressure applied to the sample. The use of a sharp razor blade is equally crucial as it mitigates mechanical damage and reduces the risk of fractures.
Enzymolysis: The primary objective of this supplementary step is to release protoplasts from tissues to facilitate observation. Although prolonged digestion could yield protoplasts, it is generally unnecessary, and the ideal approach is to minimize any disruptions.
Validation of protocol
This protocol has been used and validated in the following published articles:
Gong, J. L. et al. (2021). Illuminating the cells: transient transformation of citrus to study gene functions and organelle activities related to fruit quality. Hortic Res 8(1): e1038/s41438–021–00611–1, DOI: 10.1038/s41438-021-00611-1.
Gong, J. L. et al. (2021). Red light-induced kumquat fruit coloration is attributable to increased carotenoid metabolism regulated by FcrNAC22. Journal of experimental botany 72: 6274–6290, DOI: 10.1093/jxb/erab283.
Acknowledgments
We are grateful for the financial support from the Zhejiang Provincial Natural Science Foundation of China (LQ23C150004 and LR23C150001), the National Natural Science Foundation of China (32102318), and the Key Project for New Agricultural Cultivar Breeding in Zhejiang Province, China (2021C02066-1). This protocol was modified from our previously published work [12].
Competing interests
The authors declare no conflict of interest.
References
- Brandizzi, F., Hawes, C., Boevink, P. and Roberts, A. (2001). GFP enlightens the study of endomembrane dynamics in plant cells. Plant Biosyst. 135(1): 3–12. https://doi.org/10.1080/11263500112331350580
- Zhu, C., Zheng, X., Huang, Y., Ye, J., Chen, P., Zhang, C., Zhao, F., Xie, Z., Zhang, S., Wang, N., et al. (2019). Genome sequencing and CRISPR/Cas9 gene editing of an early flowering Mini‐Citrus (Fortunella hindsii). Plant Biotechnol. J. 17(11): 2199–2210. https://doi.org/10.1111/pbi.13132
- Sparkes, I. A., Runions, J., Kearns, A. and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1(4): 2019–2025. https://doi.org/10.1038/nprot.2006.286
- Tsuda, K., Qi, Y., Nguyen, L. V., Bethke, G., Tsuda, Y., Glazebrook, J. and Katagiri, F. (2011). An efficient Agrobacterium‐mediated transient transformation of Arabidopsis. Plant J. 69(4): 713–719. https://doi.org/10.1111/j.1365-313x.2011.04819.x
- Orzaez, D., Mirabel, S., Wieland, W. H. and Granell, A. (2006). Agroinjection of Tomato Fruits. A Tool for Rapid Functional Analysis of Transgenes Directly in Fruit. Plant Physiol. 140(1): 3–11. https://doi.org/10.1104/pp.105.068221
- Zhao, Y., Mao, W., Chen, Y., Wang, W., Dai, Z., Dou, Z., Zhang, K., Wei, L., Li, T., Zeng, B., et al. (2019). Optimization and standardization of transient expression assays for gene functional analyses in strawberry fruits. Hortic. Res. 6(1): e1038/s41438–019–0135–5. https://doi.org/10.1038/s41438-019-0135-5
- An, J. P., Wang, X. F., Li, Y. Y., Song, L. Q., Zhao, L. L., You, C. X. and Hao, Y. J. (2018). EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 Act in a Regulatory Loop That Synergistically Modulates Ethylene Biosynthesis and Anthocyanin Accumulation. Plant Physiol. 178(2): 808–823. https://doi.org/10.1104/pp.18.00068
- Li, T., Jiang, Z., Zhang, L., Tan, D., Wei, Y., Yuan, H., Li, T. and Wang, A. (2016). Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 88(5): 735–748. https://doi.org/10.1111/tpj.13289
- Shen, S. l., Yin, X. r., Zhang, B., Xie, X. l., Jiang, Q., Grierson, D. and Chen, K. s. (2016). CitAP2.10activation of the terpene synthaseCsTPS1is associated with the synthesis of (+)-valencene in ‘Newhall’ orange. J. Exp. Bot. 67(14): 4105–4115. https://doi.org/10.1093/jxb/erw189
- Yin, X., Xie, X., Xia, X., Yu, J., Ferguson, I. B., Giovannoni, J. J. and Chen, K. (2016). Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening. Plant J. 86(5): 403–412. https://doi.org/10.1111/tpj.13178
- Zhu, M., Lin, J., Ye, J., Wang, R., Yang, C., Gong, J., Liu, Y., Deng, C., Liu, P., Chen, C., et al. (2018). A comprehensive proteomic analysis of elaioplasts from citrus fruits reveals insights into elaioplast biogenesis and function. Hortic. Res. 5(1): e1038/s41438–017–0014–x. https://doi.org/10.1038/s41438-017-0014-x
- Gong, J., Tian, Z., Qu, X., Meng, Q., Guan, Y., Liu, P., Chen, C., Deng, X., Guo, W., Cheng, Y., et al. (2021). Illuminating the cells: transient transformation of citrus to study gene functions and organelle activities related to fruit quality. Hortic. Res. 8(1): e1038/s41438–021–00611–1. https://doi.org/10.1038/s41438-021-00611-1
- Smertenko, A. P., Deeks, M. J. and Hussey, P. J. (2010). Strategies of actin reorganisation in plant cells. J. Cell Sci. 123(17): 3019–3028. https://doi.org/10.1242/jcs.071126
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY license (https://creativecommons.org/licenses/by/4.0/).
How to cite
Gong, J. and Sun, X. (2024). Transient Expression Assay and Microscopic Observation in Kumquat Fruit. Bio-protocol 14(7): e4968. DOI: 10.21769/BioProtoc.4968.
Category
Plant Science > Plant cell biology > Cell imaging
Plant Science > Plant molecular biology > Protein
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









