- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In-house Extraction and Purification of Pfu-Sso7d, a High-processivity DNA Polymerase
(*contributed equally to this work) Published: Vol 14, Iss 7, Apr 5, 2024 DOI: 10.21769/BioProtoc.4967 Views: 2673
Reviewed by: Marcelo S. da SilvaRitu GuptaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2171 Views

Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2377 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 66 Views
Abstract
The polymerase chain reaction (PCR) is an extensively used technique to quickly and accurately make many copies of a specific segment of DNA. In addition to naturally existing DNA polymerases, PCR utilizes a range of genetically modified recombinant DNA polymerases, each characterized by varying levels of processivity and fidelity. Pfu-Sso7d, a fusion DNA polymerase, is obtained by the fusion of Sso7d, a small DNA-binding protein, with Pfu DNA polymerase. Pfu-Sso7d is known for its high processivity, efficiency, and fidelity but is sold at a sumptuously high price under various trade names and commercial variants. We recently reported a quick and easy purification protocol that utilizes ethanol or acetone to precipitate Pfu-Sso7d from heat-cleared lysates. We also optimized a PCR buffer solution that outperforms commercial buffers when used with Pfu-Sso7d. Here, we provide a step-by-step guide on how to purify recombinant Pfu-Sso7d. This purification protocol and the buffer system will offer researchers cost-efficient access to fusion polymerase.
Key features
• We detail a precipitation-based protocol utilizing ethanol and acetone for purifying Pfu-Sso7d.
• Despite ethanol and acetone displaying effective precipitation efficiency, acetone is preferred for its superior performance.
• Furthermore, we present a PCR buffer that outperforms commercially available PCR buffers.
• The Pfu-Sso7d purified in-house and the described PCR buffer exhibit excellent performance in PCR applications.
Background
DNA polymerases are extensively used in PCR to exponentially amplify DNA and generate a substantial quantity from a minimal initial DNA template. An efficient PCR amplification necessitates a DNA polymerase that is not only thermostable but also has excellent fidelity and processivity. These essential DNA polymerase characteristics shorten extension times and enable error-free amplification of lengthy DNA templates. A variety of approaches are utilized to enhance the processivity of the DNA polymerase. One such approach is using fusion DNA polymerases, created by the covalent fusion of a tiny DNA-binding protein to the polymerase domain of the enzyme. Pfu-Sso7d fusion DNA polymerase, for instance, is produced by the fusion of Pfu DNA polymerase with Sso7d, a tiny 7 kDa protein derived from Sulfobulus solfataricus, and binds to dsDNA in a sequence-independent manner [1,2]. This fusion significantly increases processivity by preventing the Pfu-Sso7d from frequently dissociating from the template.
Recombinant DNA polymerases are typically purified through a time-consuming, cost-intensive, two-step affinity purification followed by dialysis [3,4]. We recently reported a straightforward, economical, and time-saving method for expressing and purifying the Pfu-Sso7d fusion DNA polymerase. This involves heat denaturation and DNase I treatment of bacterial lysate to recover thermostable DNA polymerase (Figure 1). The heat-cleared and DNase I–treated lysates are then precipitated using ethanol or acetone [5]. We also reported an in-house PCR buffer system that outperforms commercially available alternatives for PCR amplification of various DNA templates. Laboratories dealing with a large number of PCRs and constrained resources can greatly benefit from the in-house purification of thermostable polymerases and the preparation of in-house buffer solutions.
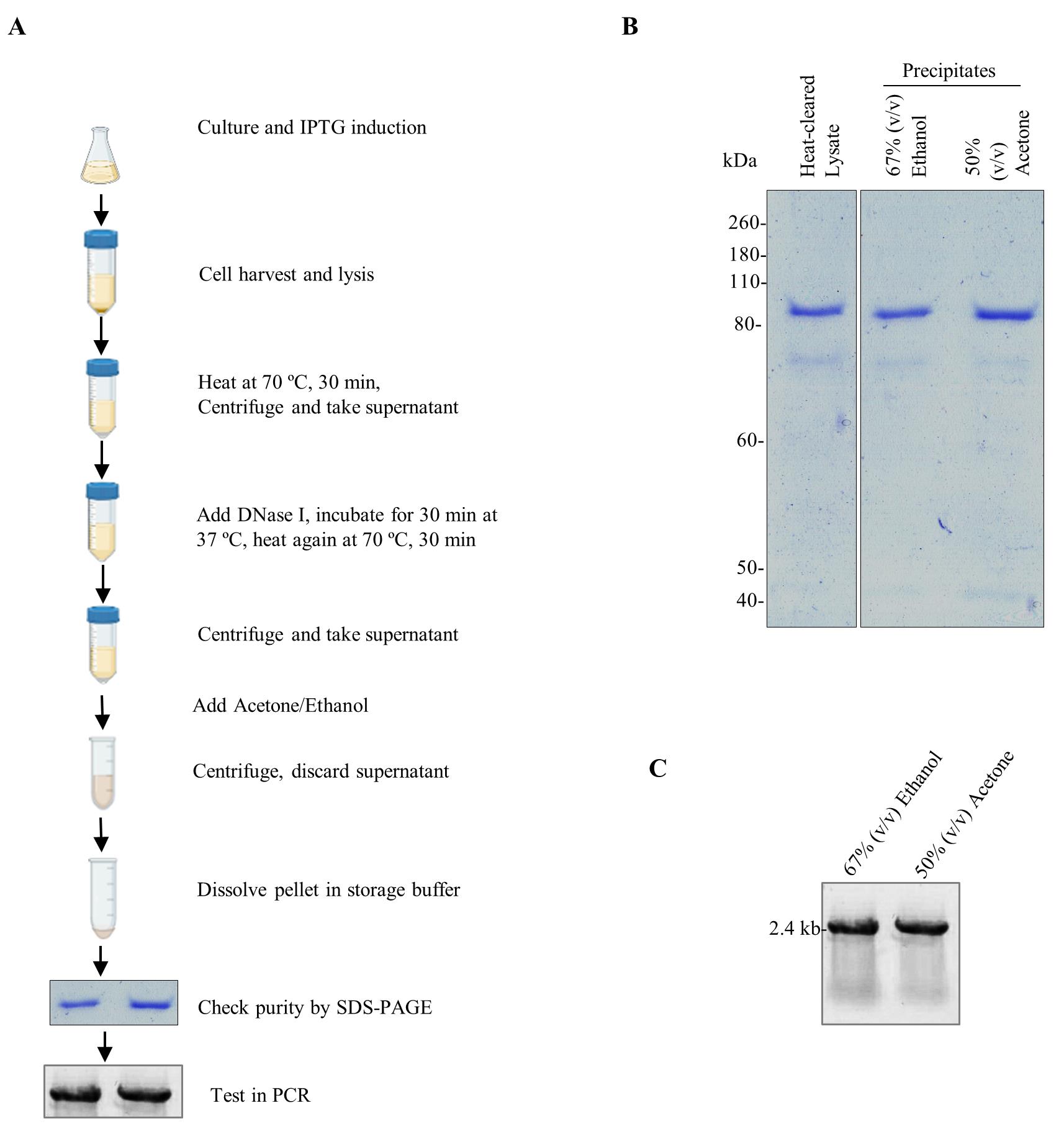
Figure 1. Precipitation-based protocol for the purification of Pfu-Sso7d fusion DNA polymerase. A. Schematic representation showing the extraction and purification of Pfu-Sso7d from the IPTG-induced bacterial culture. Briefly, a colony of BL21 (DE3) pLysS transformed with pET-28b-Pfu-Sso7d expression plasmid was cultured and induced with IPTG. Bacterial cells were harvested and lysed using a lysis buffer containing lysozyme. Heat-cleared and DNase I–treated lysates were then precipitated using acetone or ethanol and analyzed by SDS-PAGE and PCR. B. SDS-PAGE analysis of the Pfu-Sso7d in heat-cleared and DNase I–treated cell lysate and of the precipitates obtained with 67% (v/v) ethanol or 50% (v/v) acetone. C. The precipitated Pfu-Sso7d was tested in the PCR amplification of a 2.4 kb fragment from the plasmid template.
Materials and reagents
Biological materials
BL21 (DE3) pLysS cells for protein expression (Thermo Fisher Scientific, catalog number: C606010)
pET-28b-Pfu-Sso7d plasmid (a gift from Dr. Alexander Klenov, York University, Canada) (Sequence can be downloaded from https://barricklab.org/twiki/pub/Lab/ProtocolsReagentsPfuSso7d/6his-pfu-sso7d-pET28.gbk)
Reagents
Tris (hydroxymethyl) aminomethane, Tromethamine, Tris base (Sisco Research Laboratory, catalog number: 71033)
Luria broth (LB) (Himedia, catalog number: M575)
Agar (Himedia, catalog number: GRM026)
Super optimal catabolite (SOC) (Himedia, catalog number: G015)
Kanamycin (Himedia, catalog number: A008)
Chloramphenicol (Himedia, catalog number: CMS218)
Deoxyribonucleotides (dNTPs) (Sisco Research Laboratory, catalog number: 14464)
Isopropyl β-d-1-thiogalactopyranoside (IPTG) (Himedia, catalog number: MB072)
Phenylmethylsulphonyl fluoride (PMSF) (Sisco Research Laboratory, catalog number: 87606)
Lysozyme (Sisco Research Laboratory, catalog number: 45822)
Sodium dodecyl sulfate (SDS) (Sisco Research Laboratory, catalog number: 1948101)
Acrylamide 1× crystal (Sisco Research Laboratory, catalog number: 89314)
Bis-acrylamide (Sisco Research Laboratory, catalog number: 38516)
Ammonium persulfate (APS) (Himedia, catalog number: MB003)
N,N,N′,N′-Tetramethyl ethylenediamine (TEMED) (Sisco Research Laboratory, catalog number: 84666)
β-mercaptoethanol (Sisco Research Laboratory, catalog number: 83759)
Sodium phosphate monobasic (NaH2PO4) (Sigma-Aldrich, catalog number: 71505-250)
Glycerol (Sisco Research Laboratory, catalog number: 59991)
Dithiothreitol (DTT) (Sisco Research Laboratory, catalog number: 17315)
Bromophenol blue (Himedia, catalog number: GRM914)
Pair of specific primers (Integrated DNA Technology)
Deoxyribonuclease I (DNase I) (Invitrogen, catalog number: AM2222)
Agarose (Himedia, catalog number: MB002)
Betaine solution (Sigma-Aldrich, catalog number: B0300)
Ethylenediaminetetraacetic acid (EDTA) (Qualigens, catalog number: Q18455)
Ethanol, absolute 99.9% (any brand)
Acetone (Sisco Research Laboratory, catalog number: 31566)
Tween 20 (Merck, catalog number: SB3S630097)
Triton X-100 (Sisco Research Laboratory, catalog number: 64518)
Sodium chloride (NaCl) (Sisco Research Laboratory, catalog number: 76945)
Glycine (Merck, catalog number: MA7M562461)
Coomassie brilliant blue R-250 (Sisco Research Laboratory, catalog number: 93473)
Sodium hydrogen phosphate dodecahydrate (Na2HPO4·2H2O) (Sisco Research Laboratory, catalog number: 83417)
Potassium dihydrogen orthophosphate extra pure AR, 99.5% (KH2PO4) (Sisco Research Laboratory, catalog number: 50451)
Nonidet P-40 (NP-40) (Thermo Fischer Scientific, catalog number: 28324)
Bovine serum albumin solution (Sigma-Aldrich, catalog number: A8412)
Acetic acid (Sisco Research Laboratory, catalog number: 85801)
Ammonium sulphate [(NH4)2SO4] (Sisco Research Laboratory, catalog number: 88064)
Magnesium sulphate (MgSO4) (Sisco Research Laboratory, catalog number: 50014)
Potassium chloride extra pure AR, 99.5% (KCl) (Sisco Research Laboratory, catalog number: 38630)
Hydrochloric acid, 6N aqueous solution (HCl) (Sisco Research Laboratory, catalog number: 17560)
Ethidium bromide (Sigma-Aldrich, catalog number: E7637)
Magnesium chloride anhydrous extra pure, 98% (MgCl2) (Sisco Research Laboratory, catalog number: 31196)
Solutions
Phosphate buffer saline (PBS) (see Recipes)
Lysis buffer (see Recipes)
Storage buffer (see Recipes)
100 mM IPTG (see Recipes)
1 M Tris-HCl, pH 6.8 (see Recipes)
1.5 M Tris-HCl, pH 8.8 (see Recipes)
4× SDS dye (see Recipes)
10× Tris-Glycine-SDS buffer (see Recipes)
Staining solution (see Recipes)
De-staining solution (see Recipes)
10× PCR buffer (see Recipes)
Recipes
PBS (100 mL)
Reagent Final concentration Quantity Na2HPO4·2H2O 100 mM 1.779 g KH2PO4 18 mM 0.244 g NaCl 137 mM 0.8 g KCl 2.7 mM 0.02 g Adjust pH to 7.4 with HCl/NaOH Double-distilled water n/a up to 100 mL Autoclave and store at 4 °C.
Lysis buffer (50 mL)
Reagent Final concentration Quantity NaCl (5 M) 300 mM 3 mL NaH2PO4 (1 M, pH 8.0) 50 mM 2.5 mL Glycerol (100%) 10% (v/v) 5 mL Triton-X 100 (10%) 0.1% (v/v) 0.5 mL Double-distilled water n/a up to 50 mL Store at 4 °C. Add 0.5 mM PMSF and 2 mg/mL lysozyme just before use.
Storage buffer (50 mL)
Reagent Final concentration Quantity EDTA (0.5 M) 0.1 mM 0.01 mL Tris-HCl (1 M, pH 8.0) 25 mM 1.25 mL NaCl (5 M) 250 mM 2.5 mL NP-40 (100%) 0.2% (v/v) 0.1 mL Glycerol (100%) 50% (v/v) 25 mL Tween 20 (100%) 0.2% (v/v) 0.1 mL Double-distilled water n/a up to 50 mL Filter sterilize, aliquot, and store at -20 °C. Add 2 mM DTT just before use.
100 mM IPTG (10 mL)
Reagent Final concentration Quantity IPTG 100 mM 0.238 g Double-distilled water n/a up to 10 mL Filter sterilize, aliquot, and store at -20 °C.
1 M Tris-HCl, pH 6.8 (100 mL)
Reagent Final concentration Quantity Tris base 1 M 12.1 g Adjust pH to 8.8 with HCl/NaOH Double-distilled water n/a up to 100 mL Autoclave and store at 4 °C.
1.5 M Tris-HCl, pH 8.8 (100 mL)
Reagent Final concentration Quantity Tris base 1.5 M 18.1 g Adjust pH to 8.8 with HCl/NaOH Double-distilled water n/a up to 100 mL Autoclave and store at 4 °C.
4× SDS dye (5 mL)
Reagent Final concentration Quantity Tris-HCl (1 M, pH 6.8) 200 mM 1 mL Glycerol (100%) 40% (v/v) 2 mL SDS (10%) 4% (w/v) 2 mL Bromophenol blue 0.08% (w/v) 0.004 mL Double-distilled water n/a up to 5 mL Aliquot and store at -20 °C. Add β-mercaptoethanol to 5% (v/v) just before use.
10× Tris-Glycine-SDS buffer (100 mL)
Reagent Final concentration Quantity Tris base 250 mM 3.03 g SDS 35 mM 1 g Glycine 1.92 M 14.4 g Double-distilled water n/a up to 100 mL Store at room temperature.
Staining solution (500 mL)
Reagent Final concentration Quantity Methanol (100%) 40% (v/v) 200 mL Acetic acid (100%) 8% (v/v) 40 mL Coomassie brilliant blue R-250 0.1% (w/v) 0.5 g Double-distilled water n/a up to 500 mL Store at room temperature.
De-staining solution (500 mL)
Reagent Final concentration Quantity Methanol (100%) 40% (v/v) 200 mL Acetic acid (100%) 8% (v/v) 40 mL Double-distilled water n/a up to 500 mL Store at room temperature.
10× PCR buffer (25 mL)
Reagent Final concentration Quantity Tris-HCl (1.5 M, pH 8.8) 200 mM 3.3 mL KCl (1 M) 100 mM 2.5 mL (NH4)2SO4 (1 M) 100 mM 2.5 mL MgSO4 (1 M) 20 mM 0.5 mL Triton X-100 (10%) 1% (v/v) 2.5 mL Nuclease-free BSA (100 mg/mL) 1 mg/mL 0.25 mL Double-distilled water n/a up to 25 mL Filter sterilize, aliquot, and store at -20 °C.
Laboratory supplies
100 mm cell culture dishes (Sigma-Aldrich, catalog number: Z755923-150EA)
50 mL tubes (Abdos, catalog number: P10424)
1.5 mL tubes (Abdos, catalog number: P10202)
Equipment
Micropipettes 10, 100, 200, and 1,000 μL (any brand)
Orbital shaker (any brand)
Vortex mixer (Thermo Fisher Scientific, catalog number: 128101)
Water bath (MAC Serological water bath, catalog number: MSW-273)
Incubator (any brand)
Centrifuge (any brand)
Biosafety cabinet (MAC Horizontal laminar flow bench, catalog number: MSW-161)
PCR machine (Thermo Fisher Scientific, model: VeritiTM 96-well fast thermal cycler)
Agarose gel apparatus (Bio-Rad, catalog number: 1703940)
Protein electrophoresis system (Bio-Rad, model: Mini-PROTEAN® Tetra cell, catalog number: 1658005EDU)
Visible spectrophotometer (Labman, model: LMSP-V320)
Autoclave (any brand)
UV transilluminator (any brand)
Quantus fluorometer (Promega, catalog number: E6150)
Procedure
Transformation of BL21 (DE3) pLysS cells
Mix 1 μL (10–25 ng) of pET-28b-Pfu-Sso7d plasmid with 50 μL of BL21 (DE3) pLysS competent cells in a microcentrifuge tube and incubate the mixture on ice for 20–30 min.
Heat-shock the transformation tube at 42 °C for 45 s.
Put the tubes back on ice for 2 min.
Add 500 μL of prewarmed super optimal catabolite (SOC) media (without antibiotic) to the competent cells and grow at a 37 °C shaking incubator for 30–60 min.
Plate 100–200 μL of the transformation onto an LB agar plate containing kanamycin (50 μg/mL) and chloramphenicol (35 μg/mL).
Incubate the plate overnight at 37 °C.
Expression of Pfu-Sso7d in BL21 (DE3) pLysS cells
Culture a single transformed colony in 2 mL of LB media containing kanamycin (50 μg/mL) and chloramphenicol (35 μg/mL) with constant shaking at 200–250 rpm overnight at 37 °C.
Note: Glycerol stock of the culture can be stored at -80 °C.
Inoculate a 1 mL aliquot of this overnight starter culture in 100 mL of LB media containing kanamycin (50 μg/mL) and chloramphenicol (35 μg/mL) with constant shaking at 200–250 rpm at 37 °C.
When the culture's optical density at 600 nm (OD600) reaches 0.4–0.5, add 0.5 mM of IPTG and incubate the culture overnight at 18 °C with constant shaking.
Preparation of heat-cleared lysate
Harvest the bacterial cells by centrifugation at 2,000× g for 15 min at 4 °C.
Wash the cell pellet with 5 mL of PBS buffer and then resuspend it in 4.5 mL of lysis buffer freshly supplemented with 0.5 mM PMSF and 2 mg/mL lysozyme.
Incubate at 37 °C for 30 min with occasional mixing.
Spin the sample tubes briefly and then heat the tubes at 70 °C for 30 min.
Place it on ice for 15 min, centrifuge the lysate at 13,500× g for 10 min, and collect the supernatant in a fresh tube.
Add DNase I and MgCl2 to a final concentration of 40 U/mL and 2 mM, respectively.
Incubate the tubes at 37 °C for 30 min.
Spin the sample tubes briefly and heat the tubes at 70 °C for 30 min.
Centrifuge at 13,500× g for 10 min and transfer the supernatant to a fresh tube.
Save a portion of this heat-cleared lysate to check the purity and efficiency of heat denaturation on SDS-PAGE.
Acetone precipitation method
Add ice-cold acetone in a 1:1 (v/v) ratio (50% final) and keep the tubes at -20 °C for 20 min.
Centrifuge at 13,500× g for 20 min and carefully discard the supernatant. Tap the tube on a paper towel to eliminate any remaining supernatant. If required, save a portion of this supernatant to check acetone precipitation efficiency.
Add 2–3 mL of storage buffer freshly supplemented with 2 mM DTT.
Vigorously vortex the tube and leave it overnight at -20 °C.
Centrifuge the sample at 2,500× g for 5 min. Collect the supernatant, aliquot, and store at -20 °C or -80 °C.
If necessary, the pellets can be extracted once more with 1–2 mL of storage buffer and repeating steps D3 and D5.
Save a portion of this precipitated and solubilized polymerase to check the purity and acetone precipitation efficiency on SDS-PAGE.
Ethanol precipitation method
Add ethanol in 1:2 (v/v) ratios (67% final) and keep the tubes at room temperature for 20 min.
Centrifuge at 13,500× g for 20 min and carefully discard the supernatant. Tap the tube on a paper towel to eliminate any remaining supernatant. If required, save a portion of this supernatant to check ethanol precipitation efficiency.
Add 2–3 mL of storage buffer freshly supplemented with 2 mM DTT.
Vigorously vortex the tube and leave it overnight at -20 °C.
Centrifuge the sample at 2,500× g for 5 min. Collect the supernatant, aliquot, and store at -20 °C or -80 °C.
If necessary, the pellet can be extracted once more with 1–2 mL of storage buffer and repeating steps E3 and E5.
Save a portion of this precipitated and solubilized polymerase to check the purity and ethanol precipitation efficiency on SDS-PAGE.
Analysis of the purified Pfu-Sso7d
Run 10–20 μL of the various samples saved above on SDS-PAGE (10% separating gel, 5% stacking) and stain with Coomassie Brilliant Blue R-250.
To conduct PCR, combine 1× homemade or commercial PCR buffer, 200 nM primers, 0.2 mM dNTPs, 0.1–0.5 μL of polymerase, 0.7–1.5 M betaine, and an appropriate quantity of DNA template (genomic DNA: 50–250 ng; plasmid or viral DNA: 10 pg–20 ng; cDNA: up to 5 μL) in a total reaction volume of 25–50 μL for 30–35 cycles. Execute PCR with an initial denaturation at 95 °C for 30 s, followed by 25–35 cycles of denaturation at 95 °C for 10 s, primer annealing at 45–72 °C for 10–20 s, extension at 72 °C (2 kb/min), and a final extension of 5 min at 72 °C.
Subject the PCR products to electrophoresis on a 0.8%–2.0% agarose gel containing 1 μg/mL ethidium bromide and visualize the separated PCR products under UV illumination.
Fractions of acetone- and ethanol-precipitated and solubilized polymerase were diluted, and proteins were estimated using Bicinchoninic acid (BCA) method. The protein concentration was generally in the range of 1.0–4.0 μg/μL.
Validation of protocol
This protocol has been used and validated in the following research article:
Farooqui et al. [5]. Quick and easy method for extraction and purification of Pfu-Sso7d, a high processivity DNA polymerase. Protein Expression and Purification.
General notes and troubleshooting
General notes
Acetone and ethanol can both efficiently precipitate Pfu-Sso-7d, but we suggest acetone over ethanol because the former showed a relatively lesser amount of contaminating DNA and requires a lesser amount of acetone (50%) compared to ethanol (67%).
Although we have performed the heat-shock method to transform plasmid DNA into E. coli, other methods of transformation could also be followed.
When prepared and stored as advised, all the buffers are stable for up to one year; polymerases prepared through this protocol and stored as described remain stable for up to three years. We have not tested them beyond the mentioned time.
Troubleshooting
If activity loss over long-term storage at -20 °C or -80 °C is noticed, it may be recovered by supplementing Pfu-Ss07d solution with 2 mM fresh DTT.
Acknowledgments
We are grateful for the financial support from the Department of Science and Technology-Science and Engineering Research Board (SRG-2020-000819) and the University Grants Commission [F.30-564/2021 (BSR)], Government of India, through Start-Up Research Grants. The protocol is adapted from Farooqui et al. [5].
Competing interests
The authors declare no competing financial interests.
References
- Wang, Y. (2004). A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res. 32(3): 1197–1207.
- Gera, N., Hussain, M., Wright, R. C. and Rao, B. M. (2011). Highly Stable Binding Proteins Derived from the Hyperthermophilic Sso7d Scaffold. J. Mol. Biol. 409(4): 601–616.
- Dabrowski, S. and Kur, J. (1998). Recombinant His-tagged DNA polymerase. I. Cloning, purification and partial characterization of Thermus thermophilus recombinant DNA polymerase. Acta Biochim. Pol. 45(3): 653–660.
- Rivera, M., Reyes, J., Blazquez-Sanchez, P. and A Ramirez-Sarmiento, C. (2021). Recombinant protein expression and purification of codon-optimized Pfu-Sso7d v2. Doi: dx.doi.org/10.17504/protocols.io.bzusp6we.
- Farooqui, A. K., Ahmad, H., Rehmani, M. U. and Husain, A. (2023). Quick and easy method for extraction and purification of Pfu-Sso7d, a high processivity DNA polymerase. Protein Expr. Purif. 208–209: 106276.
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Mahboob, A., Fatma, N. and Husain, A. (2024). In-house Extraction and Purification of Pfu-Sso7d, a High-processivity DNA Polymerase. Bio-protocol 14(7): e4967. DOI: 10.21769/BioProtoc.4967.
Category
Biochemistry > Protein > Expression
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









