- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Mesenchymal Stromal Cell (MSC) Functional Analysis—Macrophage Activation and Polarization Assays
Published: Vol 14, Iss 6, Mar 20, 2024 DOI: 10.21769/BioProtoc.4957 Views: 4353
Reviewed by: Vivien J. Coulson-ThomasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Evaluating Human Natural Killer Cells Antibody-dependent Cellular Cytotoxicity (ADCC) Using Plate-bound Anti-CD16 Antibodies
Weiru Liu
Jan 5, 2022 4587 Views

Determination of Antibody Activity by Platelet Aggregation
Halina H.L. Leung [...] Beng H. Chong
Sep 5, 2023 2257 Views

Assessing Human Treg Suppression at Single-Cell Resolution Using Mass Cytometry
Jonas Nørskov Søndergaard [...] James B. Wing
Aug 20, 2025 2923 Views
Abstract
Stem cell–based therapies have evolved to become a key component of regenerative medicine approaches to human pathologies. Exogenous stem cell transplantation takes advantage of the potential of stem cells to self-renew, differentiate, home to sites of injury, and sufficiently evade the immune system to remain viable for the release of anti-inflammatory cytokines, chemokines, and growth factors. Common to many pathologies is the exacerbation of inflammation at the injury site by proinflammatory macrophages. An increasing body of evidence has demonstrated that mesenchymal stromal cells (MSCs) can influence the immunophenotype and function of myeloid lineage cells to promote therapeutic effects. Understanding the degree to which MSCs can modulate the phenotype of macrophages within an inflammatory environment is of interest when considering strategies for targeted cell therapies. There is a critical need for potency assays to elucidate these intercellular interactions in vitro and provide insight into potential mechanisms of action attributable to the immunomodulatory and polarizing capacities of MSCs, as well as other cells with immunomodulatory potential. However, the complexity of the responses, in terms of cell phenotypes and characteristics, timing of these interactions, and the degree to which cell contact is involved, have made the study of these interactions challenging. To provide a research tool to study the direct interactions between MSCs and macrophages, we developed a potency assay that directly co-cultures MSCs with naïve macrophages under proinflammatory conditions. Using this assay, we demonstrated changes in the macrophage secretome and phenotype, which can be used to evaluate the abilities of the cell samples to influence the cell microenvironment. These results suggest the immunomodulatory effects of MSCs on macrophages while revealing key cytokines and phenotypic changes that may inform their efficacy as potential cellular therapies.
Key features
• The protocol uses monocytes differentiated into naïve macrophages, which are loosely adherent, have a relatively homogeneous genetic background, and resemble peripheral blood mononuclear cells–derived macrophages.
• The protocol requires a plate reader and a flow cytometer with the ability to detect six fluorophores.
• The protocol provides a quantitative measurement of co-culture conditions by the addition of a fixed number of freshly thawed or culture-rescued MSCs to macrophages.
• This protocol uses assessment of the secretome and cell harvest to independently verify the nature of the interactions between macrophages and MSCs.
Graphical overview
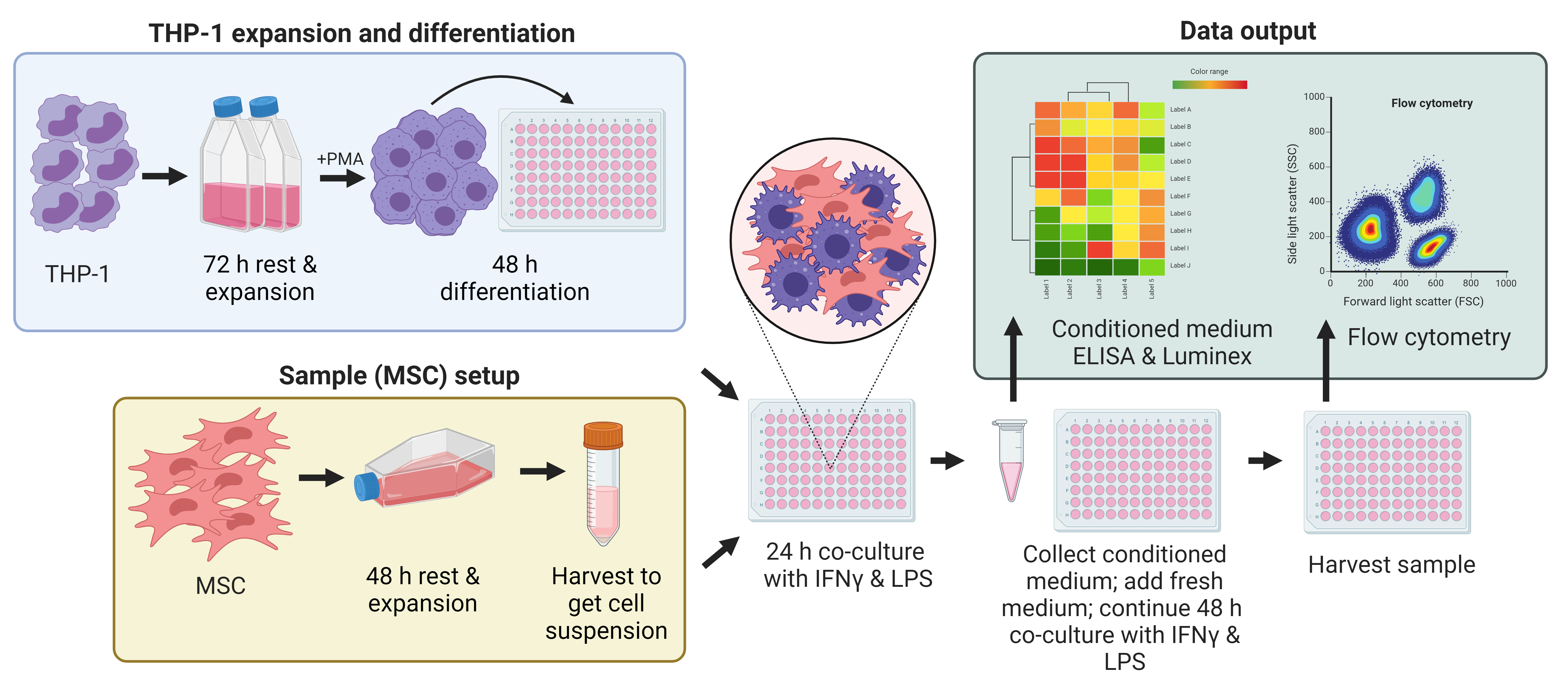
Background
Mesenchymal stromal cell (MSC) transplants, through their immunomodulatory and regenerative properties, have emerged as a viable therapeutic approach in a wide range of diseases. MSCs isolated from bone marrow, umbilical cord, and adipose tissue have been used in clinical trials to treat conditions as far-reaching as traumatic brain injury, autism spectrum disorder, osteoarthritis, immune system disorders, and cardiovascular diseases [1]. To standardize such treatments and attempt to ensure successful outcomes, various approaches have been suggested to characterize the cells in functional or potency assays in vitro, to better predict treatment responses, e.g., DOSES [donor; origin; separation method; exhibited characteristics (cell behavior); and site of delivery] [2]. A known property of MSCs is their ability to modulate the phenotype of macrophages.
Monocyte-derived macrophages are important cells in the inflammatory cascade, key to tissue healing and regeneration. In the early stages of tissue injury or throughout chronic inflammatory pathologies, macrophages are driven to polarize to an M1 state, referred to as classically activated, and will secrete a range of proinflammatory cytokines and chemokines such as tumor necrosis factor-alpha (TNFα), interleukin-12 (IL-12), and CC-chemokine receptor-7 (CCR7), and express major histocompatibility complex (MHC) II cell surface receptor (HLA-DR) [3]. As the inflammation subsides, the predominant macrophage phenotype shifts to M2 (alternative activation) and is associated with the release of anti-inflammatory cytokines such as interleukin-10 (IL-10), interleukin-13 (IL-13), and interleukin-1 receptor antagonist (IL-1RA) [4]. Polarization to M2 reparatory macrophages, effected by MSCs in situ, can result in the restoration of homeostasis and tissue repair as well as the reduction of inflammation [5].
MSCs in culture are responsive to culture conditions and secrete both free and exosome-encapsulated cytokines and microRNA. To establish the cell culture techniques that best prepare MSCs for in vivo delivery and subsequent immunomodulation of macrophage phenotype and function in the body, in vitro co-culture assays can be used to study the crosstalk between these cell types to recapitulate the complex interactions in vivo. However, studies with macrophage cultures can be challenging due to the difficulties in attaining a pure population of macrophages and in the harvesting of these cells, as they strongly adhere to tissue culture surfaces when differentiated/activated. The Tohoku Hospital Pediatrics-1 (THP-1) cell line has been widely used to study monocyte and macrophage function in vitro, in the presence of a wide range of other cell types such as intestinal cells, adipocytes, T-lymphocytes, platelets, and vascular smooth muscle cells [6].
The proposed mechanisms of action of MSCs [7–13] center on the abatement of the inflammatory condition by the reduction in a number of proinflammatory cytokines such as TNFα, IL-6, IL-8, and IL1-β. In the first part of this assay, the ability of delivered MSCs to counter this proinflammatory condition is measured by the suppression of the release of TNFα and other cytokines from M1-like macrophages. In the clinic, the mobilization of MSCs to counter inflammation, as seen with oxygen therapies administered to patients with respiratory and cognitive disorders, may mimic a similar endogenous response by MSCs [14,15].
MSC administration generates concomitant decreases in proinflammatory cytokine release and increases in anti-inflammatory mediator production by macrophages [16,17]. These changes are accompanied by the polarization of M1 towards M2 macrophages [18]. This shift is examined in the second part of the assay.
In consideration of the role of cell therapy and, in particular, the delivery of allogeneic MSCs in ameliorating disease conditions, the study of the interactions of MSCs and macrophages is pertinent. Herein, we have established an in vitro assay system to study both macrophage activation and polarization in the presence of delivered MSCs. Freshly thawed and culture-rescued cells can differ in their ability to activate and polarize macrophages. Freshly thawed cells have disturbed membrane physiology and display a differential release of paracrine mediators and microparticles (exosomes, microvesicles, and apoptotic bodies) compared with cultured cells. Although freshly thawed cells are less likely to persist in the body due to innate immune cascades, coagulation response, etc., their passive release of microparticles is likely augmented compared with culture-rescued cells. However, stronger immunomodulatory capacity has been attributed to freshly cultured cells [19]. These assays establish key parameters for studying the interaction of MSCs and macrophages in vitro.
Materials and reagents
Biological materials
THP-1 cell vial, which has been stored in liquid nitrogen (LN2) at vapor phase (ATCC, catalog number: TIB-202)
Human MSCs derived from bone marrow or umbilical cord tissue, which have been stored frozen in a cryovial and maintained in LN2 at vapor phase (RoosterBio Inc., Frederick, MD, catalog number: RoosterVial-hBM or supplied by Duke University MC3)
Reagents
RPMI 1640 containing L-glutamine and 10 mM HEPES (ATCC, catalog number: 30-2001)
RoosterNourishTM -MSC (RoosterBio Inc, catalog number: KT-001)
Prime-XV MSC Expansion XSFM (Irvine Scientific, catalog number: 91149)
Penicillin/Streptomycin (P/S) (Millipore Sigma, catalog number: P4333)
Fetal bovine serum (FBS), heat inactivated (H.I.) and characterized (Cytiva, catalog number: SH30071.01)
β-mercaptoethanol (BME) (Thermo Fisher Scientific, catalog number: 21985023)
Dimethylsulfoxide (DMSO) (Sigma-Aldrich, catalog number: C6164)
Phorbol 12-myristate 13 acetate (PMA) (Sigma-Aldrich, catalog number: P1585)
PLASMA-LYTE A (Baxter Intl Inc., catalog number: 2B2543Q)
Human serum albumin (HSA) (Albutein) 25% (Grifols, catalog number: 68516-5216-02)
Phosphate buffered saline (PBS) Ca2+/Mg2+ free, low endotoxin (Millipore Sigma, catalog number: TMS-012-A)
Interferon-gamma (IFN-γ) (Thermo Fisher Scientific, catalog number: 300-02)
Lipopolysaccharide (LPS) (Millipore Sigma, catalog number: L4391)
Sterile endotoxin-free water (VWR, catalog number: 75799-280)
TrypLE Express 1× (Millipore Sigma, catalog number: 12605028)
Zombie UV Fixable Viability kit (BioLegend, catalog number: 423107)
Human TruStain FcXTM (BioLegend, catalog number: 422302)
Antibodies (BioLegend, see Table 1)
Table 1. Antibody information for cell phenotype panel
Surface marker Final dilution Conjugate Vendor Catalog Clone Human macrophage
/MSC panel antibodiesHLA-DR 1:20 (see General note 6) FITC BioLegend 307620 L243 CD163 1:20 PE BioLegend 333606 GHI/61 CD73 1:20 PerCP/Cy5.5 BioLegend 344014 AD2 CD206 1:20 APC BioLegend 321110 15-2 CD14 1:20 PE/Cy7 BioLegend 367112 63D3 Zombie UV 1:1,000 - BioLegend 423107 - Compensation beads Anti-Mouse Ig +/- control beads One drop/500 µL FC buffer - BD 552843 - ArC reactive beads for use with Zombie UV One drop/500 µL FC buffer - Thermo Fisher A10346 - Anti-mouse negative and positive control beads (Thermo Fisher Scientific, catalog number: 552843)
ArC reactive beads (Thermo Fisher Scientific, catalog numbers: A10346 and A10628)
Cytofix fixation buffer (BD Biosciences; catalog number: 554655)
ELISA kit TNFα (e.g., Thermo Fisher Scientific, catalog number: PO1375)
Luminex Cytokine Kit Magnetic 30 Plex Panel (Thermo Scientific, catalog number: LHC6003M)
Solutions
THP-1 expansion medium (see Recipes)
THP-1 differentiation medium (see Recipes)
Diluting buffer (see Recipes)
PMA diluted (see Recipes)
LPS stock (see Recipes)
IFN-γ stock (see Recipes)
Co-culture medium (see Recipes)
Zombie-UV (see Recipes)
Blocking buffer (see Recipes)
FC buffer (see Recipes)
Antibody cocktail (see Recipes)
Recipes
THP-1 expansion medium
Reagent Final concentration Volume RPMI 1640 79% 394.5 mL FBS H.I. 20% 100 mL P/S 1% 5 mL BME 0.1% 0.5 mL Total 100% 500 mL THP-1 differentiation medium
Reagent Final concentration Volume RPMI 1640 98% 48.5 mL FBS H.I. 1% 0.5 mL P/S 1% 0.5 mL PMA diluted 100 ng/mL 0.5 mL Total 100% 50 mL Diluting buffer
Reagent Final concentration Volume PLASMA-LYTE A 96% 48 mL HSA (25%) 4% (1%) 2 mL Total 100% 50 mL PMA diluted
Make a stock solution of 5 mg of PMA in 1 mL of DMSO. Then, add 5 µL of stock to 2.5 mL of RPMI 1640 (no additives). Filter sterilize through a 0.2 µm filter and then add to 1% FBS media as above.
Reagent Final concentration Volume PMA stock (5 mg/mL in DMSO) 10 µg/mL 5 µL Medium RPMI 1640 2.5 mL Total 100% 2.5 mL LPS stock
Reagent Final concentration Quantity LPS 1 mg/mL 1 mg Sterile water 1 mL Total 100% 1 mL IFN-γ stock
Reagent Final concentration Quantity IFN-γ 100 µg/mL 100 µg Sterile water 1 mL Total 100% 1 mL Co-culture medium
Reagent Final concentration Volume RPMI 1640 98% 49 mL FBS H.I. 1% 0.5 mL P/S 1% 0.5 mL LPS 100 ng/mL 5 µL IFN-γ 100 ng/mL 50 µL Total 100% 50 mL Zombie- UV
Reagent Final concentration Volume Zombie-UV (reconstituted) 0.1% 5 µL PBS Ca2+/Mg2+ free (1×) 99.9% 5 mL Total 100% 5 mL Blocking buffer
Reagent Final concentration Volume TruStain FcX 10% 1 mL FC buffer 90% 9 mL Total 100% 10 mL FC buffer
Reagent Final concentration Volume FBS H.I. 2% 2 mL PBS Ca2+/Mg2+ free (1×) 98% 98 mL Total 100% 100 mL Antibody cocktail (70 wells)
Reagent Final concentration Volume Antibodies (350 µL each) 50% 1.75 mL FC buffer 50% 1.75 mL Total 100% 3.5 mL
Laboratory supplies
Non-treated T-25, T-75, and T-175 suspension flasks (e.g., Corning, ultralow attachment, catalog number: 4616, 3814)
Tissue culture treated T-225 flasks (e.g., Corning, CELLBIND, catalog number: 3293)
Cryogenic vials for vapor phase (e.g., Corning, Fisher Scientific, catalog number: 431386)
Freezing container (e.g., Corning, Millipore Sigma, model: CoolCell LX, catalog number: CLS432002)
Serological pipettes [e.g., VWR, catalog numbers: 414004-266 (5 mL); 414004-267 (10 mL); 414004-268 (25 mL)]
Micropipette tips, P10P1000 µL [e.g., Fisher Scientific, Biotix uTIP filter, catalog numbers: 12-111-000 (10 µL); 12-111-004 (300 µL); 12-111-132 (1,000 µL)]
Via-1-CassetteTM cartridges (Chemometec, catalog number: 941-0012)
CELLBIND 96-well cell culture plate (Corning, Millipore Sigma, catalog number: CLS3300)
96-well culture plate for media storage (e.g., Corning, tissue culture treated, catalog number: 3894)
V-bottom 96-well plate (Corning, Millipore Sigma, catalog number: CLS3894)
Parafilm (e.g., Sigma-Aldrich, catalog number: P7793)
Pre-saturated alcohol wipes (e.g., VWR, catalog number: 33503-136)
Equipment
Biosafety cabinet (BSC) level A2 (e.g., Thermo Fisher Scientific, catalog number: 1377)
Incubator set to 5% CO2 and 37 °C (e.g., Thermo Fisher Scientific, model: HeraCell VIOS)
Cell counter, (e.g., Chemometec, model: NucleoCounter NC-200TM)
Refrigerated centrifuge capable of reaching 500× g and using 50 mL conical tubes and microplates (e.g., Thermo Fisher Scientific, model: Megafuge 8R, catalog number: 75007214)
Micropipettes P10–1,000 µL (e.g., Eppendorf, Research Plus range, catalog number: 312300020)
Pipette controller (e.g., Eppendorf, model: Easypet, catalog number: 2231000955)
Inverted microscope (e.g., Thermo Fisher Scientific, model EVOS M5000, catalog number: AMF5000)
Microplate absorbance reader (e.g., Cytation 5)
Flow cytometer, e.g., CytoFlex (Beckman Coulter), LSRFortessa (BD)
Luminescence microplate reader (e.g., Bio-Rad, model: Bioplex 200)
Software and datasets
Flow cytometry software is needed for data analysis. This can be either FlowJoTM (BD Biosciences) or CytoBank (Beckman Coulter) software
Prism 9 (GraphPad, San Diego, CA)
JMP statistical software (Pro 17)
Procedure
Establishment of macrophage cultures
Thaw a vial of THP-1 cells (7 × 106 cells). Place the cell vial in a foam float in a water bath at 37 °C and time for 2 min. Check that 90% of the ice has disappeared before removing it from the water bath.
Wipe the vial with a pre-saturated alcohol wipe. Add the cells using a 1,000 µL micropipette and tip to 9 mL of THP-1 expansion medium that has been prewarmed to 37 °C in a 50 mL conical tube and resuspend the cells using a 10 mL serological pipette.
Count the cells by taking up 60 µL of cell suspension into a Via-1 Cassette and placing the cassette into the NucleoCounter. Record the cell count and cell viability.
Centrifuge the cells at 300× g for 5 min at 4 °C. Aspirate the supernatant and add 30 mL of expansion medium to the pellet. Resuspend the pellet using a 10 mL serological pipette.
Add 30 mL of fresh prewarmed expansion medium to a T-175 suspension culture flask and add the 30 mL of cell suspension to give 60 mL total. This will seed cells at a cell density of 1.2 × 105/mL.
Incubate the flasks at 37 °C and 5% CO2 in an incubator. Feed the cell cultures twice a week by removing all media from the flask into 2 × 50 mL conical tubes using a 25 mL serological pipette, centrifuging the tubes at 300× g for 5 min at 4 °C, and resuspending each cell pellet in 30 mL of fresh media. Use a total of 60 mL of media that has been prewarmed to 37 °C for each media change.
Check the cell density at each feeding by taking up 60 µL of cell suspension in a Via-1 cassette as above. Once the cultures have reached 1 × 106/mL, they can be further divided as below (see Figure 1 and Troubleshooting 1 and 2).
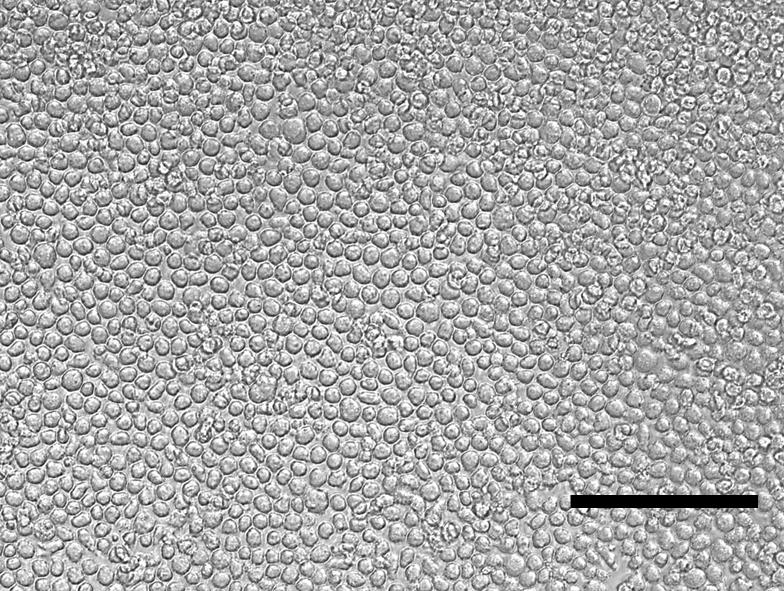
Figure 1. THP-1M loosely adherent cells after 48 h of phorbol 12-myristate 13 acetate (PMA) treatment (20×). Scale bar = 150 µm.Split the THP-1 cell suspension 1:5 into T-175 suspension culture flasks (with 60 mL of media in each) and further culture the cells to 1 × 106/mL.
Harvest the THP-1 cells by removing all the media from the flasks using a 25 mL serological pipette, centrifuging to obtain pellets, and resuspending the pellets in 1 mL of cryopreservation medium comprising 10% DMSO: 90% FBS H.I. at 10 M cells/vial. The vials can be cryopreserved by placing them in a freezing container at -80 °C overnight before storing in vapor phase LN2.
Five days before the start of the co-culture (day -5), thaw THP-1 cells in the expansion medium as above.
Centrifuge at 300× g for 5 min at 4 °C.
Count cells. Resuspend THP-1 cells in 6 mL of expansion media at 8 × 105 /mL in T-25 suspension flasks, standing upright to restrict the surface area available. Incubate for 72 h at 37 °C and 5% CO2.
On day -2, harvest THP-1 cells from T-25 flasks. Count the cells to check viability (> 90%) and cell size (~13–14 µm).
Centrifuge at 300× g for 5 min at 4 °C and resuspend the THP-1 cells at 5 × 105/mL in differentiation media (PMA treatment).
Seed 200 µL of cell suspension in each well (105/well) of a CELLBIND 96-well flat-bottom plate.
Note: Set up extra wells for the THP-1 macrophage without MSCs (THP-1M) and the fluorescence minus one (FMO) controls.
Incubate at 37 °C and 5% CO2 for 48 h to generate naïve macrophages (now termed THP-1M for macrophages).
Preparation of MSC cultures
On the same day of PMA treatment (day -2), thaw the MSCs in diluting buffer or the media of choice (RoosterNourish/XSFM) for the culture-rescued (CR) group.
Thaw one vial of cells of each MSC donor in a water bath for 2 min. Then, add the cells from the vial to 9 mL of diluting buffer or media prewarmed to 37 °C and resuspend using a 10 mL serological pipette.
Count as before and seed MSC at 2,000 (XSFM) or 3,700 (RoosterNourish) cells/cm2 in T-225 flasks with MSC growth medium. The volume of media required is typically 40–42 mL.
Note: Required MSC seeding density may vary by manufacturer/supplier. Follow the supplier recommendations.
Incubate at 37 °C and 5% CO2 for 48 h.
On day 0, harvest the MSC by washing the flask with 10 mL of PBS Ca2+/Mg2+ free, aspirating the PBS, and adding 10 mL of TrypLE solution.
Incubate the flask for 5 min at 37 °C to allow for cell detachment. Check under a microscope to see that all the cells are detached from the surface of the flask.
Transfer the cell suspension to a tube and wash the flask with MSC growth medium. Count the MSC.
Centrifuge at 500× g for 10 min at 4 °C and resuspend CR MSC at 2.5 × 106/mL or 1.25 × 106/mL in diluting buffer/media of choice.
On the day of MSC harvest (day 0), thaw extra MSC in diluting buffer or media of choice for the out-of-thaw (OOT) group.
Centrifuge at 500× g for 10 min at 4 °C and resuspend OOT MSC at 2.5 × 106/mL or 1.25 × 106/mL in diluting buffer/media of choice (see General note 1).
Establishment of co-cultures
On day 0, centrifuge the naïve macrophages plate at 300× g for 5 min at 4 °C. Gently aspirate PMA media without disturbing the cells.
Add 40 µL of MSC cell suspension (CR or OOT) into each well, resulting in MSC:THP-1M of 1:1 or 1:2 (1 × 105 MSC:1 × 105 macrophages or 0.5 × 105 MSC: 1 × 105 macrophages). Set up extra wells containing both cell types for FMOs for flow staining (see General note 2).
Prepare co-culture medium by adding LPS and IFNγ to 1% FBS RPMI 1640, prewarmed to 37 °C.
Caution: Take care to avoid skin exposure to LPS.
Add 200 µL of co-culture medium to THP-1 M-only wells. Add 160 µL of co-culture medium to wells containing MSCs.
Critical: Mix by triturating the cells three times to ensure uniform distribution of MSCs and macrophages. The macrophage plate will have the loosely adherent cells distributed on one side of the well due to centrifugation.
Incubate the co-culture plate at 37 °C and 5% CO2 for 24 h.
Removal and storage of conditioned media
On day 1, bring the co-culture plate to the BSC but do not centrifuge.
Collect 60 µL of supernatant from each well (not FMO) and place into the corresponding wells of another 96-well plate so that the plate map is the same. Collect another 40 µL supernatant from each well and place into a second 96-well plate. Secure the plate with a plate sealer and parafilm. Store the plates at -20 °C for Luminex and ELISA assays.
Add 100 µL of fresh co-culture medium containing LPS and IFNγ (see General note 3).
Incubate co-culture plate at 37 °C and 5% CO2 for a further 48 h.
Harvest of cells after co-culture
On day 3, aspirate 100 µL of co-culture supernatant from each well and discard (see General note 4).
Triturate the remaining supernatant and move to a 96-well V-bottom plate.
Wash wells once with 100 µL of PBS and transfer PBS to the V-bottom plate following the plate map.
Centrifuge the V-bottom plate at 500× g for 5 min at 4 °C and decant the supernatant. Decanting is best performed by immediately removing the plate from the centrifuge after spinning and removing the plate lid with one hand while flicking the plate liquid into the sink/biohazard container with the other hand in a swift but firm movement. The plate is then righted, and the lid replaced. The pellets should remain intact at the bottom of the plate.
Add 50 µL of TrypLE to each well in the co-culture plate and incubate at 37 °C for 5 min to detach the cells (see General note 5).
Collect and transfer cell suspension to the V-bottom plate.
Use 50 µL of fresh co-culture medium (w/o LPS or IFNγ) to wash each well and transfer the medium to the V-bottom plate.
Repeat step E7 for an additional wash.
Centrifuge the V-bottom plate at 500× g for 5 min at 4 °C and decant the supernatant.
Add 200 µL of PBS to sample wells in the V-bottom plate. Add 50 µL of PBS to FMO wells, pool all samples together in a microcentrifuge tube, and then divide between eight wells for FMO and unstained samples.
Flow staining for macrophage phenotype
Centrifuge the V-bottom plate at 500× g for 5 min at 4 °C and decant the supernatant.
Prepare Zombie-UV live-dead dye in PBS (see Recipes 8).
Add 100 µL of Zombie UV to each of the cell pellets except unstained and Zombie FMO.
Add 100 µL of PBS to unstained and Zombie FMO.
Resuspend the cells in the staining solution in each well.
Incubate in the dark for 15 min at room temperature (RT).
Centrifuge the V-bottom plate at 500× g for 5 min at 4 °C and decant the supernatant.
Wash each well with 200 µL of FC buffer.
Centrifuge the V-bottom plate at 500× g for 5 min at 4 °C and decant the supernatant.
Prepare Fc block staining solution. Add 50 µL of Fc block staining solution to all wells and resuspend three times (see Recipes 9).
Incubate in the dark for 5 min at RT.
Prepare the antibody cocktail using FC buffer (see Recipes 10) plus 10% v/v of HLA-DR, CD163, CD73, CD206, and CD14 (see Table 1 for dilutions, conjugates, and catalog numbers).
Without removing Fc block, add 50 µL of antibody cocktail to each of the sample wells.
Treat the FMOs by adding 5 µL of each antibody except the one listed (FITC FMO has no FITC antibody).
Add 50 µL of FC buffer to the unstained well. Resuspend all wells three times.
Incubate in the dark for 20 min at 4 °C.
Add 100 µL of FC buffer for washing, centrifuge the V-bottom plate at 500× g for 5 min at 4 °C, and decant the supernatant.
Repeat step F17 for an additional wash.
Add 100 µL of CytoFix fixation buffer to the pellets and resuspend three times. Incubate in the dark for 15 min at 4 °C.
Add 100 µL of FC buffer to each well to wash, centrifuge the V-bottom plate at 500× g for 5 min at 4 °C, and decant the supernatant.
Resuspend the cell pellet in 100 µL of FC buffer. Keep the plate at 4 °C and analyze using a flow cytometer within three days. If the flow cytometer does not have a high throughput system for plates, transfer the samples to the appropriate tubes for your system prior to flow.
Prepare compensation beads for the antibody panel by adding one drop of positive and one drop of negative anti-mouse beads (see Table 1) to each of the six tubes and adding 1–5 µL of single antibody into each single stain tube, leaving one as unstained. Vortex for 5 s and then incubate in the dark at RT for 15–30 min. Add 2 mL of FC buffer to wash. Centrifuge at 500× g for 5 min. Carefully remove the supernatant and resuspend in 400 µL of FC buffer.
Prepare compensation beads for Zombie staining by using the ArC reactive beads in place of the anti-mouse beads and only incubate Zombie dye with the positive beads (component A). Once washed as above, add the negative beads into the tube. Use all beads for flow compensation.
Analysis of growth factors, cytokines, and chemokines in conditioned media
Thaw conditioned media plates and use aliquots for the TNFα ELISA and the Luminex assay (see manufacturers’ instructions).
Data analysis
The results for the TNFα ELISA comprise absorbance values, which are converted to concentrations of TNFα via the standard curve generated in the assay. It is recommended to set up six replicate wells for THP-1M only and each sample group of MSCs and THP-1M (biological replicates). The results are normalized to the TNFα release from macrophages in the absence of MSCs and are calculated as % inhibition of the THP-1M response. Data are presented as mean ± standard error of the mean. Statistical analysis was performed using Prism 9 (GraphPad, San Diego, CA) using one-way analysis of variance (ANOVA) with post-hoc Tukey’s tests on paired comparisons.
The results for the Luminex 30 Plex Panel comprise fluorescence intensity measurements, which are converted to concentrations based on the standard curves generated for the 30 analytes. It is recommended to set up four replicate wells for THP-1M only and each sample group of MSCs and THP-1M (biological replicates). Readings are considered outliers if they fall outside of the linear portion of the standard curves. The concentrations can be standardized using JMP statistical software (Pro 17) to give a Z-score heatmap.
Since the polarization assay is downstream of the activation assay, the same number of biological replicates (n = 6) is used for each MSC donor. The polarization assay generates flow data, which can be compensated on the instrument (using single stains) and gated using FMOs and unstained samples using FlowJo or CytoBank software. Once populations have been established, the data are presented as HLA-DR+ cells as a percentage of CD14+ live cells (M1) or CD206+/CD14+ (single positive M2), CD163+/CD14+ (single positive M2), or CD206+/CD163+/CD14+ (double positive M2) cells.
Validation of protocol
Macrophage activation assay
Previous work in which the activation assay was described and validated is found in Medrano-Trochez et al. ([20], Figures 6 and 7), and Pradhan et al. ([21], Figures 2 and 6).
Evidence that inhibition of TNFα has occurred can be sought by a relatively straightforward ELISA. Once that has been determined, the conditioned media can then be subjected to a Luminex for comparison of multiple analytes. Two THP-1 cell vials from different culture batches (B1 = batch 1 and B2 = batch 2) were used in this assay. The profiles of the THP-1 B1 and B2 are similar. The protocol is robust and reproducible since each co-culture is compared to its THP-1M-only control. The activation assay was validated using bone marrow–derived and umbilical cord tissue–derived MSCs [21,20] (Pradhan et al., 2020; Medrano-Trochez et al., 2021). Separate data on three MSC donors are shown in Figure 2. Overall, the culture-rescued cells showed less inhibition of the TNFα release from macrophages than freshly thawed MSCs.
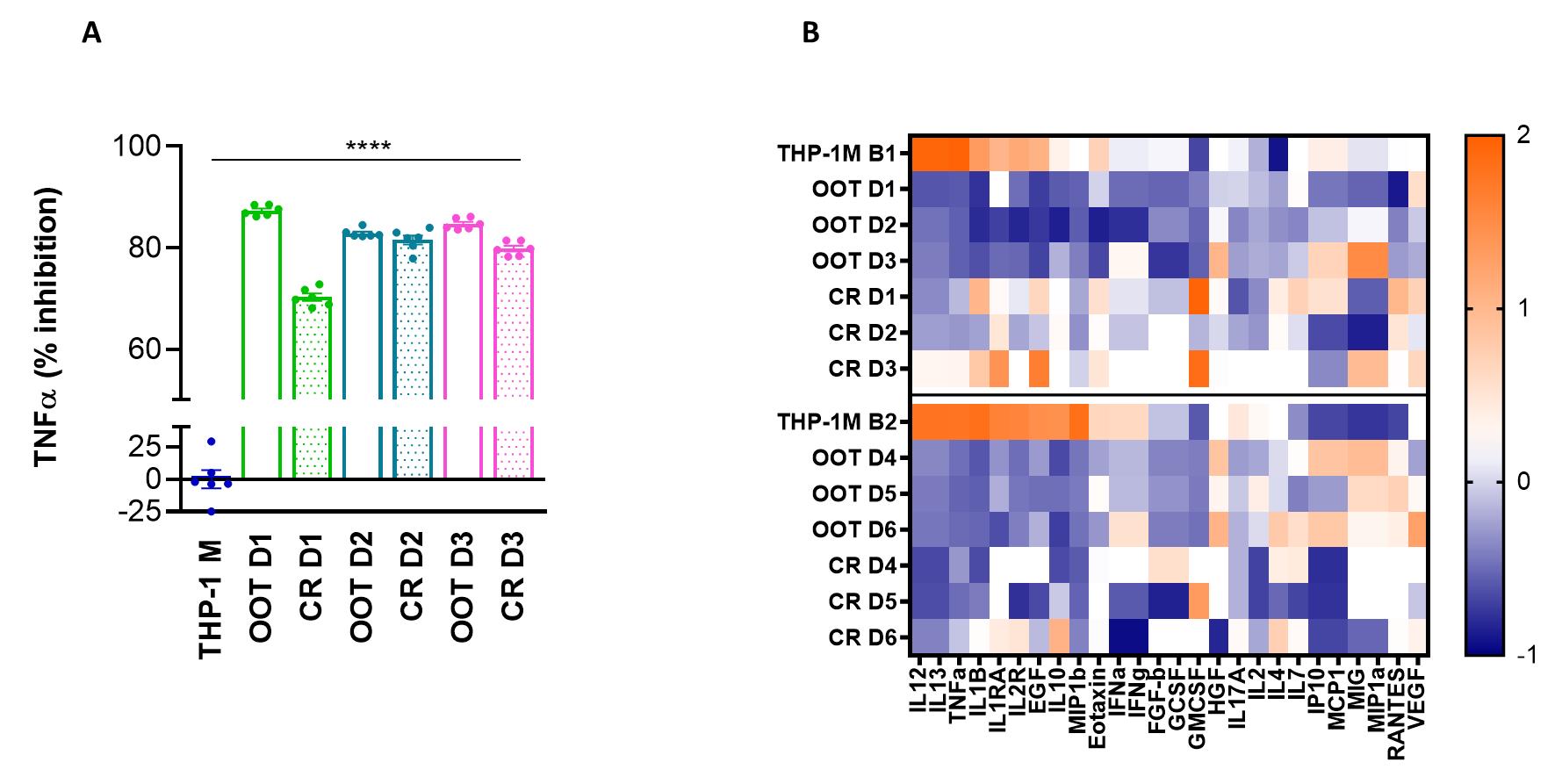
Figure 2. Percent inhibition of tumor necrosis factor-alpha (TNFα) by mesenchymal stromal cells (MSC) donor and Z-score of analytes in conditioned media. A. Values of TNFα concentration in the media are measured using ELISA and used as a baseline to calculate the % inhibition of the response when MSCs are added. THP-1M is macrophage only; OOT = out-of-thaw/freshly thawed; CR = culture rescued. All donor samples are significantly different from THP-1M. B. For six donors OOT and CR, a panel of analytes present in the co-culture media is presented as Z scores, where orange is high, and blue is low concentration.
Macrophage polarization assay
Phenotypic analysis of cells was performed at harvest by flow cytometry. The percentage of cells expressing surface markers outlined in the methods section was analyzed following an initial gating strategy of forward and side scatter > singlets > live cells (Zombie UV live/dead discrimination) on > 30K acquired events. In wells containing MSCs, there was a high surface marker expression of CD73, and this was used to gate out MSCs. The cell population that expressed CD14 was then identified and used to measure levels of HLA-DR for THP-1M and THP-1M and MSC donor. The gating strategy is shown in detail in Figure 3 as cell plots and in Figure 4 as gating hierarchy. This protocol was validated using bone marrow–derived and umbilical cord tissue–derived MSCs for both out-of-thaw and culture-rescued cells. Data for five donors, added at a ratio of 1:2 (MSCs: THP-1M), are shown in Figure 5. Overall, MSCs showing a greater loss of HLADR positivity showed a larger extent of polarization to M2, at least for CD163 positivity.
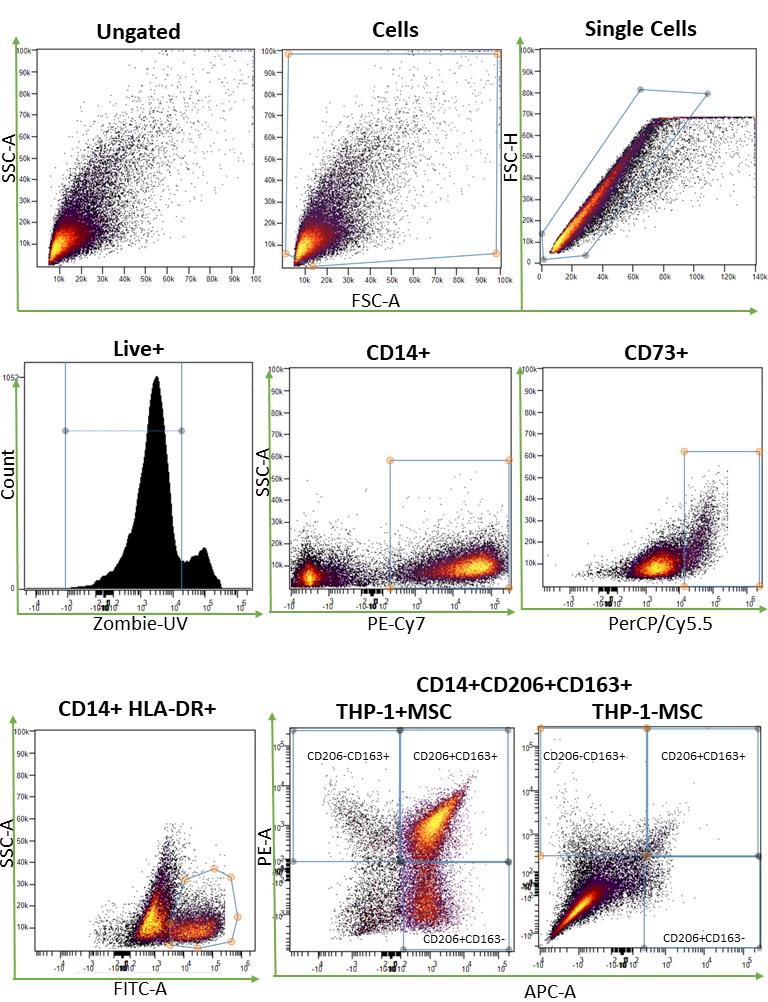
Figure 3. Illustration of gating steps needed to establish macrophage phenotype (HLA-DR+, CD163+/CD14+, CD206+/CD14+, or CD163+/CD206+/CD14+ cells) and presence of mesenchymal stromal cells (MSCs) (CD73+). Gating strategy shown in the sequence of panels from CytoBank.
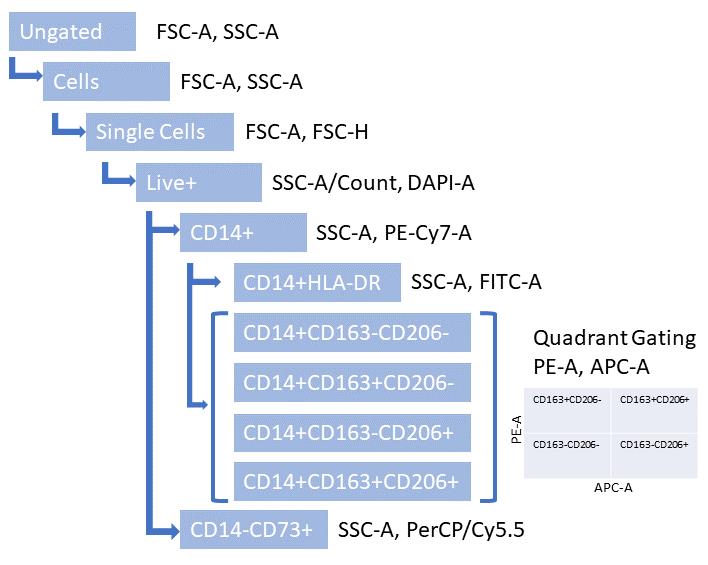
Figure 4. Illustration of gating hierarchy for separation of cell populations. The final gate is a quadrant for CD163 and CD206 marker analysis.
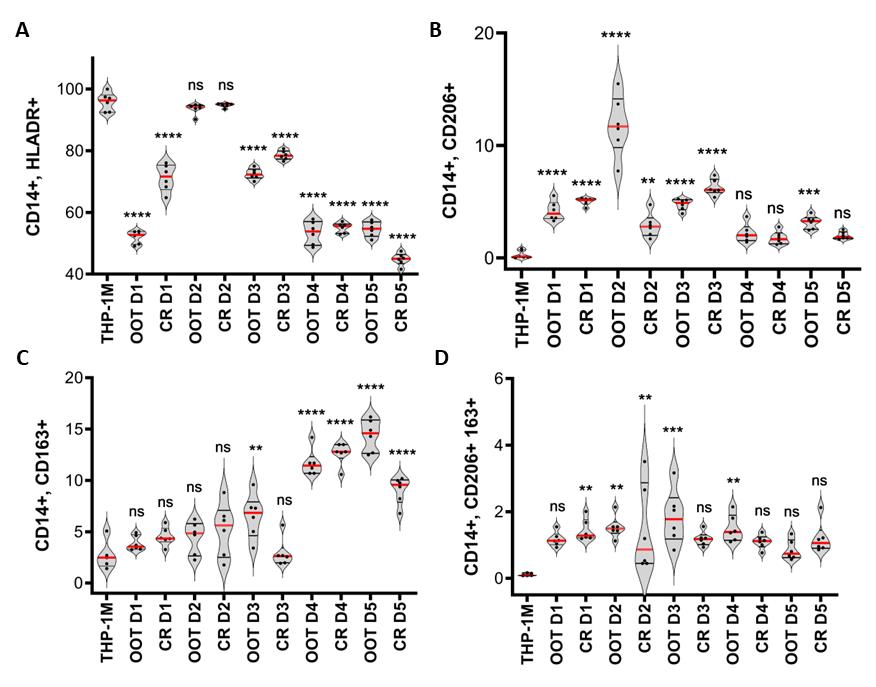
Figure 5. Macrophage phenotype determined at harvest by flow cytometry. Macrophage phenotype determined for THP-1M and with mesenchymal stromal cells (MSC) in co-culture at harvest. A. HLA-DR positive (M1-like) macrophages as a percentage of CD14+ live cells, measured with and without MSCs, with all groups showing significant M1 marker reduction apart from donor 2. B. CD206 positive (M2-like) macrophages as a percentage of CD14+ live cells. C. CD163 positive (M2-like) macrophages as a percentage of CD14+ live cells. Donors 4 and 5 showed lower HLA-DR positivity. D. Double positive CD206/CD163 (M2-like) macrophages as a percentage of CD14+ live cells. The statistical test applied was a one-way ANOVA with Tukey’s post-hoc test; *(p < 0.05), **(p < 0.001), ***(p < 0.001), ****(p < 0.0001); * denotes significance compared to THP-1M group.
General notes and troubleshooting
General notes
MSCs can be delivered in diluting buffer or media. Both have been used successfully for this protocol. Importantly, the same delivery vehicle should be in all MSC samples, with the same cell concentration and the same volume for comparisons to be made.
This protocol demonstrates the assay for bone marrow–derived MSCs and umbilical cord tissue–derived MSCs. Additional optimization may be required for thaw, culture, and harvest if using MSCs derived from another tissue source as well as ratios of MSCs to THP-1M.
In our hands, the maintenance of the THP-1M M1 state required the sustained presence of LPS and IFNγ in the culture media. The release of TNF-α, coupled with the establishment of M1 phenotype, can be used to optimize the concentration of LPS needed in the assay.
A successful harvest from the co-culture plate requires a two-stage process. The loosely adherent macrophages will be in the media at the bottom of the well, so removing media without disturbing the lowest layer is important. After collecting the macrophages in a wash step, enzyme digestion is needed to detach the MSCs from the plate.
This step can be avoided if the phenotype of the THP-1Ms is the only cell type under investigation. The TrypLE can be used if MSCs are required in the flow analysis.
As 50 µL of Fc block is left in each well before the antibody cocktail is added, the final dilution is 1/20 for 5 µL of antibody. It is recommended to first titrate antibody concentrations to test detection limits for particular cytometers. A concentration of as little as 1 µL of antibody per well (1:100), for each of the five-panel antibodies, has been demonstrated to work well in this assay.
Troubleshooting
The method to culture THP-1 monocytes has been described before [22] and has been modified to deal with the many issues encountered with expanding and maintaining these cells [23]. The cells are freezer sensitive and can become exhausted if expanded too much or less proliferative if not dense enough. A rule of thumb is to observe the diameter of the cells. Healthy viable cells are 13–14 µm and dead/dying cells are 6–7 µm, determined using a cell counter. Once cell size decreased, they became irretrievable in our hands. Once expanded, however, they can provide a very consistent cell line for functional assays.
If THP-1 monocytes are slowing down in proliferation during the expansion phase, add 1:1 conditioned media to fresh media at each feeding.
Acknowledgments
This work was supported by the Billi and Bernie Marcus Foundation and the National Science Foundation Engineering Research Center for Cell Manufacturing Technologies (NSF EEC 1648035). Previous work in which the activation assay was described and validated is found in Medrano-Trochez et al. [20] and Pradhan et al. [21]. Graphical overview created with BioRender.com.
Competing interests
The authors have no competing interests that may influence the material presented in this protocol.
Ethical considerations
No human subjects were used for any experiments included in this study. All procedures using MSCs derived from umbilical cord tissue were supplied by collaborators at Duke University in accordance with the ethical standards and approved by the ethics committee of the institutional review boards at Georgia Institute of Technology and Duke University (IRB Protocol No. H17348).
References
- Chetty, S., Yarani, R., Swaminathan, G., Primavera, R., Regmi, S., Rai, S., Zhong, J., Ganguly, A. and Thakor, A. S. (2022). Umbilical cord mesenchymal stromal cells—from bench to bedside. Front. Cell Dev. Biol. 10: e1006295.
- Galderisi, U., Peluso, G. and Di Bernardo, G. (2021). Clinical Trials Based on Mesenchymal Stromal Cells are Exponentially Increasing: Where are We in Recent Years? Stem Cell Rev. Rep. 18(1): 23–36.
- Labonte, A. C., Tosello-Trampont, A. C. and Hahn, Y. S. (2014). The Role of Macrophage Polarization in Infectious and Inflammatory Diseases. Mol. Cells 37(4): 275–285.
- Gibon, E., Loi, F., Córdova, L. A., Pajarinen, J., Lin, T., Lu, L., Nabeshima, A., Yao, Z. and Goodman, S. B. (2016). Aging Affects Bone Marrow Macrophage Polarization: Relevance to Bone Healing. Regener. Eng. Transl. Med. 2(2): 98–104.
- Harrell, C. R., Djonov, V. and Volarevic, V. (2021). The Cross-Talk between Mesenchymal Stem Cells and Immune Cells in Tissue Repair and Regeneration. Int. J. Mol. Sci. 22(5): 2472.
- Chanput, W., Mes, J. J. and Wichers, H. J. (2014). THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 23(1): 37–45.
- Mei, S. H. J., Haitsma, J. J., Dos Santos, C. C., Deng, Y., Lai, P. F. H., Slutsky, A. S., Liles, W. C. and Stewart, D. J. (2010). Mesenchymal Stem Cells Reduce Inflammation while Enhancing Bacterial Clearance and Improving Survival in Sepsis. Am. J. Respir. Crit. Care. Med. 182(8): 1047–1057.
- Gupta, N., Su, X., Popov, B., Lee, J. W., Serikov, V. and Matthay, M. A. (2007). Intrapulmonary Delivery of Bone Marrow-Derived Mesenchymal Stem Cells Improves Survival and Attenuates Endotoxin-Induced Acute Lung Injury in Mice. J. Immunol. 179(3): 1855–1863.
- Pedrazza, L., Cunha, A. A., Luft, C., Nunes, N. K., Schimitz, F., Gassen, R. B., Breda, R. V., Donadio, M. V. F., de Souza Wyse, A. T., Pitrez, P. M. C., et al. (2017). Mesenchymal stem cells improves survival in LPS‐induced acute lung injury acting through inhibition of NETs formation. J. Cell. Physiol. 232(12): 3552–3564.
- Jackson, M. V., Morrison, T. J., Doherty, D. F., McAuley, D. F., Matthay, M. A., Kissenpfennig, A., O'Kane, C. M. and Krasnodembskaya, A. D. (2016). Mitochondrial Transfer via Tunneling Nanotubes is an Important Mechanism by Which Mesenchymal Stem Cells Enhance Macrophage Phagocytosis in the In Vitro and In Vivo Models of ARDS. Stem Cells 34(8): 2210–2223.
- Johnson, C. L., Soeder, Y. and Dahlke, M. H. (2016). Mesenchymal stromal cells for immunoregulation after liver transplantation. Curr. Opin. Organ Transplant. 21(6): 541–549.
- Min, H., Xu, L., Parrott, R., Overall, C. C., Lillich, M., Rabjohns, E. M., Rampersad, R. R., Tarrant, T. K., Meadows, N., Fernandez-Castaneda, A., et al. (2020). Mesenchymal stromal cells reprogram monocytes and macrophages with processing bodies. Stem Cells 39(1): 115–128.
- Stevens, H. Y., Bowles, A. C., Yeago, C. and Roy, K. (2020). Molecular Crosstalk Between Macrophages and Mesenchymal Stromal Cells. Front. Cell Dev. Biol. 8: e600160.
- Patry, C., Doniga, T., Lenz, F., Viergutz, T., Weiss, C., Tönshoff, B., Kalenka, A., Yard, B., Krebs, J., Schaible, T., et al. (2020). Increased mobilization of mesenchymal stem cells in patients with acute respiratory distress syndrome undergoing extracorporeal membrane oxygenation. PLoS One 15(1): e0227460.
- Cozene, B., Sadanandan, N., Farooq, J., Kingsbury, C., Park, Y. J., Wang, Z. J., Moscatello, A., Saft, M., Cho, J., Gonzales-Portillo, B., et al. (2021). Mesenchymal Stem Cell-Induced Anti-Neuroinflammation Against Traumatic Brain Injury. Cell Transplant. 30: 096368972110357.
- Behnke, J., Kremer, S., Shahzad, T., Chao, C. M., Böttcher-Friebertshäuser, E., Morty, R. E., Bellusci, S. and Ehrhardt, H. (2020). MSC Based Therapies—New Perspectives for the Injured Lung. J. Clin. Med. 9(3): 682.
- Liu, X., Fang, Q. and Kim, H. (2016). Preclinical Studies of Mesenchymal Stem Cell (MSC) Administration in Chronic Obstructive Pulmonary Disease (COPD): A Systematic Review and Meta-Analysis. PLoS One 11(6): e0157099.
- Gu, X., Xu, J., Yang, X., Peterson, E. and Harding, P. (2015). Fractalkine neutralization improves cardiac function after myocardial infarction. Exp. Physiol. 100(7): 805–817.
- Cottle, C., Porter, A. P., Lipat, A., Turner-Lyles, C., Nguyen, J., Moll, G. and Chinnadurai, R. (2022). Impact of Cryopreservation and Freeze-Thawing on Therapeutic Properties of Mesenchymal Stromal/Stem Cells and Other Common Cellular Therapeutics. Curr. Stem Cell Rep. 8(2): 72–92.
- Medrano-Trochez, C., Chatterjee, P., Pradhan, P., Stevens, H. Y., Ogle, M. E., Botchwey, E. A., Kurtzberg, J., Yeago, C., Gibson, G., Roy, K., et al. (2021). Single-cell RNA-seq of out-of-thaw mesenchymal stromal cells shows tissue-of-origin differences and inter-donor cell-cycle variations. Stem Cell Res Ther 12(1): 565.
- Pradhan, P., Chatterjee, P., Stevens, H. Y., Glen, C., Medrano-Trochez, C., Jimenez, A., Kippner, L., Seeto, W. J., Li, Y., Gibson, G., et al. (2020). Single-Cell Transcriptomic Attributes and Unbiased Computational Modeling for the Prediction of Immunomodulatory Potency of Mesenchymal Stromal Cells. bioRxiv 2020.09.12.294850.
- Pietilä, M., Lehtonen, S., Tuovinen, E., Lähteenmäki, K., Laitinen, S., Leskelä, H. V., Nätynki, A., Pesälä, J., Nordström, K., Lehenkari, P., et al. (2012). CD200 Positive Human Mesenchymal Stem Cells Suppress TNF-Alpha Secretion from CD200 Receptor Positive Macrophage-Like Cells. PLoS One 7(2): e31671.
- Lyons, C. (2017). Mastering the Art of Growing THP-1 cells Retrieved from https://bitesizebio.com/31538/mastering-art-growing-thp-1-cells/
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Stevens, H. Y., Jimenez, A. C., Wang, B., Li, Y., Selvam, S. and Bowles-Welch, A. C. (2024). Mesenchymal Stromal Cell (MSC) Functional Analysis—Macrophage Activation and Polarization Assays. Bio-protocol 14(6): e4957. DOI: 10.21769/BioProtoc.4957.
Category
Immunology > Immune mechanisms > In vitro model
Cell Biology > Cell-based analysis > Inflammatory response
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










