- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
SUrface SEnsing of Translation (SUnSET), a Method Based on Western Blot Assessing Protein Synthesis Rates in vitro
Published: Vol 14, Iss 3, Feb 5, 2024 DOI: 10.21769/BioProtoc.4933 Views: 5046
Reviewed by: Chiara AmbrogioMayank GautamRupkatha Banerjee

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Multi-phase Training in Virus-Like Particle Synthesis to Foster Science Self-efficacy in Students With Minimal Laboratory Experience
Macie A. Proctor-Roser [...] Naomi R. Lee
Jul 20, 2025 1734 Views
An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
Saurabh Upadhyay
Aug 20, 2025 2374 Views

Artificial Metalloenzymes in Artificial Sanctuaries Through Liquid–Liquid Phase Separation
Kaixin Wang [...] Tong Wu
Oct 5, 2025 1536 Views
Abstract
As the most energy- and metabolite-consuming process, protein synthesis is under the control of several intrinsic and extrinsic factors that determine its fine-tuning to the cellular microenvironment. Consequently, variations in protein synthesis rates occur under various physiological and pathological conditions, enabling an adaptive response by the ce•ll. For example, global protein synthesis increases upon mitogenic factors to support biomass generation and cell proliferation, while exposure to low concentrations of oxygen or nutrients require translational repression and reprogramming to avoid energy depletion and cell death. To assess fluctuations in protein synthesis rates, radioactive isotopes or radiolabeled amino acids are often used. Although highly sensitive, these techniques involve the use of potentially toxic radioactive compounds and require specific materials and processes for the use and disposal of these molecules. The development of alternative, non-radioactive methods that can be easily and safely implemented in laboratories has therefore been encouraged to avoid handling radioactivity. In this context, the SUrface SEnsing of Translation (SUnSET) method, based on the classical western blot technique, was developed by Schmidt et al. in 2009. The SUnSET is nowadays recognized as a simple alternative to radioactive methods assessing protein synthesis rates.
Key features
• As a structural analogue of aminoacyl-transfer RNA, puromycin incorporates into the elongating peptide chain.
• Detection of puromycin-labeled peptides by western blotting reflects translation rates without the need for radioactive isotopes.
• The protocol described here for in vitro applications is derived from the SUnSET method originally published by Schmidt et al. (2009).
Background
As a structural analogue of aminoacyl-tRNA, the aminonucleoside antibiotic puromycin is incorporated through non-hydrolysable peptide bounding into the growing peptide chain along the elongation process (Nathans, 1964). While high concentrations block the elongation phase and hence translation, at low doses the overall translation rate of protein synthesis remains unchanged. Consequently, the rate of formation of puromycin-labeled peptides mirrors the rate of protein translation. Taking advantage of this property, 3H-puromycin labeling was first used in 1979 to assess the rate of protein synthesis in various tissues in vivo under nutrient- and protein-poor diets (Nakano and Hara, 1979). The SUnSET (SUrface SEnsing of Translation) technique was developed 30 years later by Schmidt and colleagues to detect variations in protein synthesis rates in cultured cells by western blotting (Schmidt et al., 2009). This method was then coupled with other techniques (fluorescence-activated cell sorting or immunohistochemistry) to assess protein synthesis at different scales. Further developments, notably based on the Clik-it technology combined with O-propargyl puromycin (OP-Puro), a puromycin analogue, enable visualization of puromycilated proteins and assessment of elongation rates in tissues (Morral et al., 2020). The main advantage of using puromycin and its analogues is that it does not require radioactive isotopes such as 35S-methionine, historically used to measure protein synthesis rates, with comparable analytical performance (Schmidt et al., 2009). Because proteostasis defects are associated with a variety of chronic diseases (cancer, tissue fibrosis, inflammatory syndromes, etc.) or aging, it is necessary to monitor the changes in protein synthesis rates in response to a variety of stressors in order to better understand the translational reprogramming underlying cell adaptation. The following protocol describes a SUnSET method (Figure 1) suitable for assessing protein synthesis rates in lysates from cells grown in vitro by conventional western blot analysis.
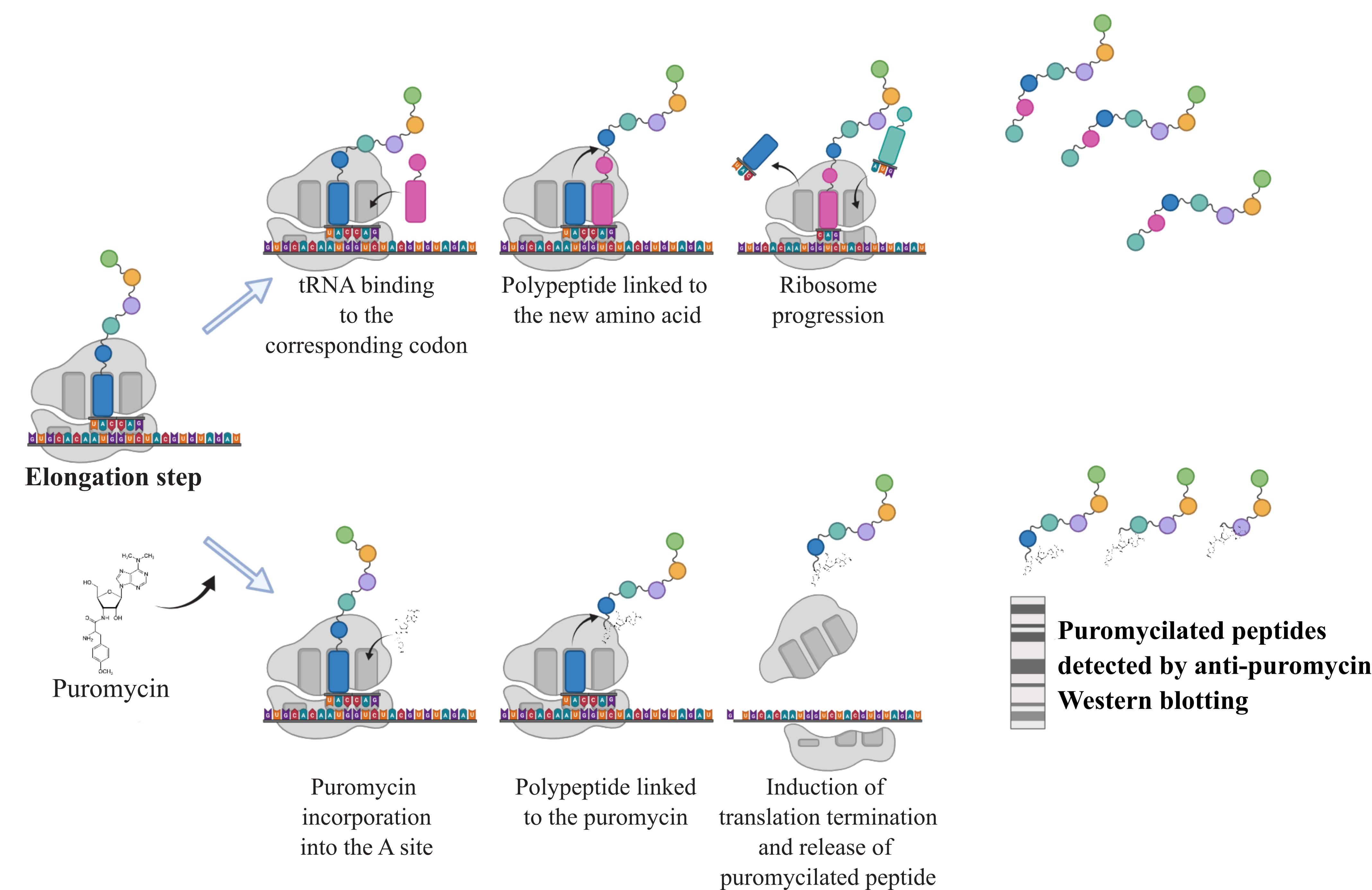
Figure 1. Principle of the SUnSET assay. During the elongation step, addition of puromycin leads to its incorporation into the A site of the ribosome. The transfer and linkage of the polypeptide to the puromycin cause the termination of translation releasing puromycilated proteins that can subsequently be detected by western blotting against the puromycin.
Materials and reagents
Biological materials
Cell lines of interest obtained from the American Type Culture Collection (ATCC). In this study, we used the colon cancer cell line HCT116.
Reagents
Puromycin (Sigma-Aldrich, catalog number: P9620)
PBS (Sigma-Aldrich, catalog number: D1408)
Complete anti-protease (Roche, catalog number: 11836145001)
Dry milk (Régilait, catalog number: 304934416704)
Bovine serum albumin (BSA) (Roche, catalog number: 10735094001)
DC Protein Assay kit (Bio-Rad, catalog number: 5000111)
Prestained protein ladder (Euromedex, catalog number: 06P-0111)
Immobilion Forte western substrate (Merck Millipore, catalog number: WBLUF0500)
Ponceau red solution (Sigma-Aldrich, catalog number: 141194)
Mouse anti-puromycin antibody (clone 12D10) (Merck Millipore, catalog number: MABE343)
Mouse anti-tubulin antibody (clone DM1A) (Sigma, catalog number: T6199)
HRP-conjugated anti-mouse secondary antibody (Cell Signaling Technology, catalog number: 7076)
Stripping buffer (Thermo Scientific, catalog number: 21059)
Solutions
RIPA protein lysis buffer 2× (see Recipes)
Tris-buffered saline-Tween (TBS-T) (see Recipes)
TBS-T 5% BSA (see Recipes)
TBS-T 5% dry milk (see Recipes)
Laemmli 6× (see Recipes)
Recipes
RIPA protein lysis buffer 2×
Reagent Final concentration Tris-HCl pH 7.2 100 mM NaCl 300 mM EDTA 10 mM Sodium deoxycholate 2% SDS 20% 0.1% Triton 100× 2× Na3VO4 4 mM β glycerophosphate 20 mM NaF 20 mM Complete anti-protease 2× Dilute to 1× in H2O.
Tris-buffered saline-Tween (TBS-T)
Reagent Final concentration Tris-HCl pH 7.5 50 mM NaCl
Tween
150 mM
0.1%
Add 5% of BSA or dry milk for obtaining TBS-T 5% BSA or 5% dry milk, respectively.
Laemmli 6×
Reagent Final concentration Tris-HCl pH 6.8 0.5 M Glycerol 1% SDS 20% 2% DTT 0.6 M Bromophenol blue 0.4%
Laboratory supplies
6-well or 10 cm diameter tissue culture plates
Sterile scrapers
Microcentrifuge tubes
Plastic wrap
Equipment
Tissue culture apparatus (tissue culture hood, CO2 incubator, etc.)
Pipettes and micropipettes
Vacuum pump
Centrifuge (4 °C)
Cold room
Heat block
Western blotting apparatus (SDS-PAGE running cassette, power supply, shaker, transfer cassette, nitrocellulose membrane, etc.)
ChemiDoc imaging system (Bio-Rad, catalog number: 12003153)
Software and datasets
Fiji (National Institutes of Health)
Procedure
Puromycin incorporation in vitro
Seed HTC116 cells one day before the experiment and maintain at 37 °C and 5% CO2 to allow attachment to the tissue culture plate. In the experiment presented in Figure 2, 200,000 cells per well were seeded in a 6-well plate.
On the day of the experiment, apply the studied treatment on cells for the desired time.
Before harvesting and 15 min before the end of the treatment, expose cells to 5 μg/mL puromycin directly diluted in the media. All samples must be incubated with the same concentration of puromycin for an equal period.
Protein extracts
At the end of the 15-min incubation with puromycin, remove media and rinse cells once with cold PBS.
Put the tissue culture plate on ice and incubate with 1× RIPA protein lysis buffer (see Recipes) containing proteases and phosphatases inhibitors (1 volume of 1× RIPA for 1 volume of cell pellet) for 20 min.
Scrape the cells and collect in a microcentrifuge tube.
Centrifuge at 13,000× g for 20 min at 4 °C to get rid of cellular debris.
Collect the supernatant in a new microcentrifuge tube and add the appropriate volume of Laemmli 6× (see Recipes).
Dose the amounts of extracted proteins using the DC Protein Assay kit according to manufacturer’s instructions.
Add the appropriate volume of Laemmli 1× to normalize protein concentrations in all samples and denaturate by heating at 95 °C for 5 min.
Western blotting
Load 20 μg of proteins for all studied conditions on a 10% SDS-PAGE gel. We recommend adding 5 μL of protein ladder to estimate the molecular weight of visualized proteins.
When separated, transfer proteins onto nitrocellulose membranes as a standard western blot protocol. Stain with Ponceau red solution to check equal protein amount loading before electrophoresis.
Block the membrane with TBS-T 5% dry milk for 1 h at room temperature with gentle shaking.
Wash with TBS-T for 5 min on a shaker and repeat the operation two times.
Incubate overnight at 4 °C on a gentle shaker with puromycin antibody diluted at 1/10,000 into TBS-T 5% BSA.
Wash with TBS-T for 5 min on a shaker and repeat the operation two times.
Incubate at room temperature for 1 h with the HRP-conjugated anti-mouse secondary antibody (1/10,000 dilution) into TBS-T 5% dry milk.
Wash with TBS-T for 5 min on a shaker and repeat the operation two times.
Gently dry the membrane using paper towels and place it face up and flat on a sheet of plastic wrap.
Directly add the Immobilion Forte western substrate onto the whole membrane.
Detect chemiluminescence with the ChemiDoc imaging system (Figure 2). Intensity of the smear depends on cell ability to incorporate puromycin and is thus representative of protein synthesis rate.
Rinse the membrane in TBS-T and transfer in 5 mL of stripping buffer.
Protect from the light and put on thorough agitation for 15 min. Check that all bound antibodies have been detached by verifying that no chemiluminescent signal is detected on the ChemiDoc imaging system.
Block the membrane with TBS-T 5% dry milk for 30 min at room temperature on gentle shaking.
Incubate at room temperature for 1 h with the anti-tubulin antibody diluted into TBS-T 5% dry milk.
Wash with TBS-T for 5 min on a shaker and repeat the operation two times.
Incubate at room temperature for 1 h with the HRP-conjugated anti-mouse secondary antibody (1/10,000 dilution) into TBS-T 5% dry milk.
Wash with TBS-T for 5 minutes on a shaker and repeat the operation two times.
Gently dry the membrane using paper towels and place it face up and flat on a sheet of plastic wrap.
Directly add 1 mL of the Immobilion Forte western substrate in order to cover the whole membrane.
Detect chemiluminescence with the ChemiDoc imaging system (Figure 2). Intensity of the bands is proportional to protein loading before electrophoresis.
Quantification of the puromycin and tubulin signals can be performed using the Fiji software (refer to Data analysis section).
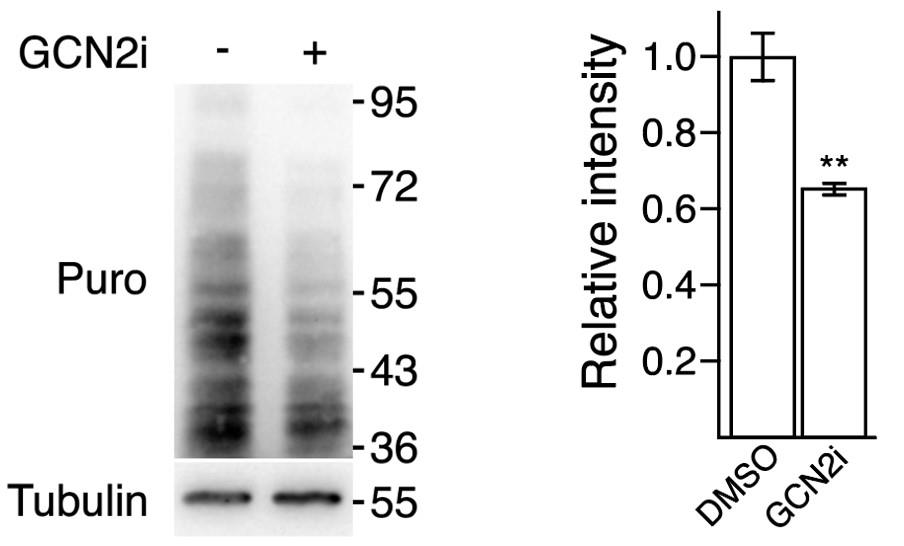
Figure 2. SUnSET assay demonstrating a repression of protein synthesis upon treatment with GCN2 kinase inhibitor. HCT116 cells were treated or not for 24 h with a GCN2 kinase inhibitor (GCN2i also named TAP20) and exposed to puromycin (5 μg/mL) 15 min before protein extraction. Protein translation rates were assessed by western blot analysis using an anti-puromycin antibody (Puro). Tubulin is presented here as a loading control. The corresponding molecular weights are indicated on the right of the western blots. Results extracted from Piecyk et al. (2023). Quantification of the data on the left represents the mean ± SEM (n = 3). Unpaired two-tailed t-test with p-value (** p < 0.01).
Data analysis
Open the Fiji software.
Click on the File menu to seek for the ChemiDoc images of the SUnSET experiment.
Use the Rectangular tool to graph a frame around the first smear to quantify and define the region of interest.
In the Analyze menu, select Gels and Select First Lane.
Move the section to the next lane and select Gels and Select Next Lane in the Analyze menu.
Repeat the operation for each lane.
Select Plot Lanes in the Analyze/Gels menu to create each lane profile plot.
Define a closed area for each lane plot using the Straight Line Selection Tool to draw a baseline for each peak.
Measure each peak area clicking inside with the Wand tool.
Report area measurements in a Results sheet.
Repeat this process for tubulin signal quantification.
Normalize: for each lane, calculate the ratio SUnSET area/Tubulin area.
Compare the ratio observed in each lane to assess the impact of studied conditions on protein synthesis rate.
Validation of protocol
This protocol was adapted from the original article: Schmidt et al. (2009). The SUnSET method was performed and validated for in vitro applications in several articles from Dr. Chaveroux’s group (Sarcinelli et al., 2020; Piecyk et al., 2021 and 2023) and other teams in the literature (e.g., Martineau et al., 2014; Mesclon et al., 2017; Arioka et al., 2020; Fong et al., 2021). The SUnSET principle has been adapted for in vivo and ex vivo applications (Goodman et al., 2011; Morral et al., 2020) and at the single-cell level coupled to flow cytometry for energy metabolism assessment by Dr. Pierre’s group (Argüello et al., 2020).
Acknowledgments
This work was supported by the Cancéropôle CLARA (CVPPRCAN000174, CVPPRCAB000180 and CV-2021-039), Region Auvergne Rhone-Alpes (19-010898-01), Institut National Du Cancer (PLBIO22-227), Projets Fondation and Aide doctorale (R16173CC, ARCMD-Doc22021020003295) from ARC, Ligue Nationale contre le Cancer (R17167CC, R19007CC), Institut Convergence François Rabelais (17IA66ANR-PLASCAN-MEHLEN), and the IPR (Innovation Pharmaceutique et Recherche) program. Figure 1 was created with BioRender.com. This protocol was adapted from the original article: Schmidt et al., 2009.
Competing interests
The authors declare no competing interests.
References
- Argüello, R. J., Combes, A. J., Char, R., Gigan, J.-P., Baaziz, A. I., Bousiquot, E., Camosseto, V., Samad, B., Tsui, J., Yan, P., et al. (2020). SCENITH: A Flow Cytometry-Based Method to Functionally Profile Energy Metabolism with Single-Cell Resolution. Cell Metab. 32(6): 1063–1075.e7. https://doi.org/10.1016/j.cmet.2020.11.007
- Arioka, Y., Shishido, E., Kushima, I., Suzuki, T., Saito, R., Aiba, A., Mori, D. and Ozaki, N. (2020). Chromosome 22q11.2 deletion causes PERK-dependent vulnerability in dopaminergic neurons. EBioMedicine 63: 103138. https://doi.org/10.1016/j.ebiom.2020.103138
- Fong, M. Y., Yan, W., Ghassemian, M., Wu, X., Zhou, X., Cao, M., Jiang, L., Wang, J., Liu, X., Zhang, J., et al. (2021). Cancer‐secreted miRNAs regulate amino‐acid‐induced mTORC1 signaling and fibroblast protein synthesis. EMBO Rep. 22(2): e51239. https://doi.org/10.15252/embr.202051239
- Goodman, C. A., Mabrey, D. M., Frey, J. W., Miu, M. H., Schmidt, E. K., Pierre, P. and Hornberger, T. A. (2011). Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 25(3): 1028–1039. https://doi.org/10.1096/fj.10-168799
- Martineau, Y., Azar, R., Müller, D., Lasfargues, C., El Khawand, S., Anesia, R., Pelletier, J., Bousquet, C. and Pyronnet, S. (2014). Pancreatic tumours escape from translational control through 4E-BP1 loss. Oncogene 33(11): Article 11. https://doi.org/10.1038/onc.2013.100
- Mesclon, F., Lambert-Langlais, S., Carraro, V., Parry, L., Hainault, I., Jousse, C., Maurin, A.-C., Bruhat, A., Fafournoux, P. and Averous, J. (2017). Decreased ATF4 expression as a mechanism of acquired resistance to long-term amino acid limitation in cancer cells. Oncotarget 8(16): 27440–27453. https://doi.org/10.18632/oncotarget.15828
- Morral, C., Stanisavljevic, J., Hernando-Momblona, X., Mereu, E., Àlvarez-Varela, A., Cortina, C., Stork, D., Slebe, F., Turon, G., Whissell, G., et al. (2020). Zonation of Ribosomal DNA Transcription Defines a Stem Cell Hierarchy in Colorectal Cancer. Cell Stem Cell 26(6): 845–861.e12. https://doi.org/10.1016/j.stem.2020.04.012
- Nakano, K. and Hara, H. (1979). Measurement of the protein-synthetic activity in vivo of various tissues in rats by using [3H]Puromycin. Biochem. J. 184(3): 663–668. https://doi.org/10.1042/bj1840663
- Nathans, D. (1964). PUROMYCIN INHIBITION OF PROTEIN SYNTHESIS: INCORPORATION OF PUROMYCIN INTO PEPTIDE CHAINS. Proc. Natl. Acad. Sci. U.S.A. 51(4): 585–592. https://doi.org/10.1073/pnas.51.4.585
- Piecyk, M., Triki, M., Laval, P.-A., Dragic, H., Cussonneau, L., Fauvre, J., Duret, C., Aznar, N., Renno, T., Manié, S. N., et al. (2021). Pemetrexed Hinders Translation Inhibition upon Low Glucose in Non-Small Cell Lung Cancer Cells. Metabolites 11(4): 198. https://doi.org/10.3390/metabo11040198
- Piecyk, M., Triki, M., Laval, P.-A., Duret, C., Fauvre, J., Cussonneau, L., Machon, C., Guitton, J., Rama, N., Gibert, B., et al. (2023). The stress sensor GCN2 differentially controls ribosome biogenesis in colon cancer according to the nutritional context. Mol. Oncol.. https://doi.org/10.1002/1878-0261.13491
- Sarcinelli, C., Dragic, H., Piecyk, M., Barbet, V., Duret, C., Barthelaix, A., Ferraro-Peyret, C., Fauvre, J., Renno, T., Chaveroux, C., et al. (2020). ATF4-Dependent NRF2 Transcriptional Regulation Promotes Antioxidant Protection during Endoplasmic Reticulum Stress. Cancers (Basel) 12(3): 569. https://doi.org/10.3390/cancers12030569
- Schmidt, E. K., Clavarino, G., Ceppi, M. and Pierre, P. (2009). SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6(4): Article 4. https://doi.org/10.1038/nmeth.1314
Article Information
Copyright
© 2024 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Piecyk, M., Fauvre, J., Duret, C., Chaveroux, C. and Ferraro-Peyret, C. (2024). SUrface SEnsing of Translation (SUnSET), a Method Based on Western Blot Assessing Protein Synthesis Rates in vitro. Bio-protocol 14(3): e4933. DOI: 10.21769/BioProtoc.4933.
Category
Biochemistry > Protein > Synthesis
Molecular Biology > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









