- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Published: Vol 13, Iss 23, Dec 5, 2023 DOI: 10.21769/BioProtoc.4895 Views: 1741
Reviewed by: Chhuttan L MeenaKarem A CourtAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Assay for Protealysin-like Protease Inhibitor Activity
Igor M. Berdyshev [...] Ilya V. Demidyuk
Oct 5, 2022 2153 Views
Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1561 Views
Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1896 Views
Abstract
Bio-hydrogen production is an eco-friendly alternative to commercial H2 production, taking advantage of natural systems. Microbial hydrogenases play a main role in biological mechanisms, catalyzing proton reduction to molecular hydrogen (H2) formation under ambient conditions. Direct determination is an important approach to screen bacteria with active hydrogenase and accurately quantify the amount of H2 production. Here, we present a detailed protocol for determining hydrogenase activity based on H2 production using methyl viologen (MV2+) as an artificial reductant, directly monitored by gas chromatography. Recombinant Escherichia coli is used as a hydrogenase-enriched model in this study. Even so, this protocol can be applied to determine hydrogenase activity in all biological samples.
Key features
• This protocol is optimized for a wide variety of biological samples; both purified hydrogenase (in vitro) and intracellular hydrogenase (in vivo) systems.
• Direct, quantitative, and accurate method to detect the amount of H2 by gas chromatography with reproducibility.
• Requires only 2 h to complete and allows testing various conditions simultaneously.
• Kinetic plot of H2 production allows to analyze kinetic parameters and estimate the efficiency of hydrogenase from different organisms.
Graphical overview
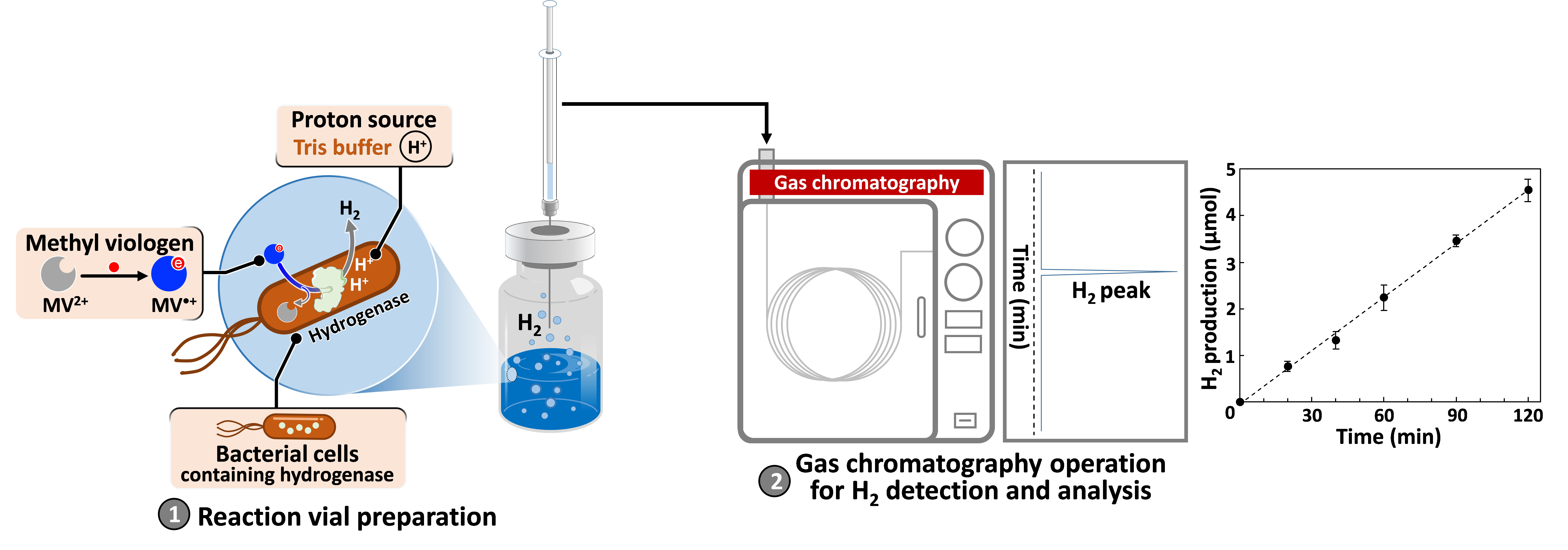
Background
Although hydrogen (H2) has been considered as a clean energy carrier, most of today’s commercial H2 production is from fossil fuels, which emit greenhouse gases into the atmosphere. Finding a more sustainable alternative remains a challenge. It is well known that, for over a million years of evolution, microorganisms have acted as a natural H2 production plant through the action of enzymes, namely the hydrogenase family (Tard and Pickett, 2009). With efficient biochemical mechanisms, it is possible to develop an environmentally friendly process for H2 production at ambient conditions by taking advantage of hydrogenase-expressing microorganisms. Among the enzymes with different catalytic sites, [FeFe]-hydrogenase is more efficient than [NiFe]- and [Fe]-types in H2 production with NADH as a reductant in a natural mechanism, as shown in Figure S1 in the Supporting materials (Ogo et al., 2020; Xuan et al., 2023). To screen microorganisms carrying active hydrogenase in a laboratory scale, methyl viologen (MV2+), which is compatible with many enzymes (Orgill et al., 2015), can also be used as an artificial reductant in this reaction. According to the theory, a more negative redox potential of methyl viologen [E(MV2+/MV•+) = -0.446 V vs. normal hydrogen electrode (NHE)] provides a favorable potential scale for proton reduction [E(H+/H2) = -0.41 V vs. NHE] at physiological pH.
In this protocol, a reduced methyl viologen (MV•+) is formed in sodium dithionite (Na2S2O4) solution to serve as an artificial chemical reductant. With cell permeability, MV•+ can penetrate and transfer electrons to intracellular hydrogenase (Kosem et al., 2023), as shown in Figure 1.
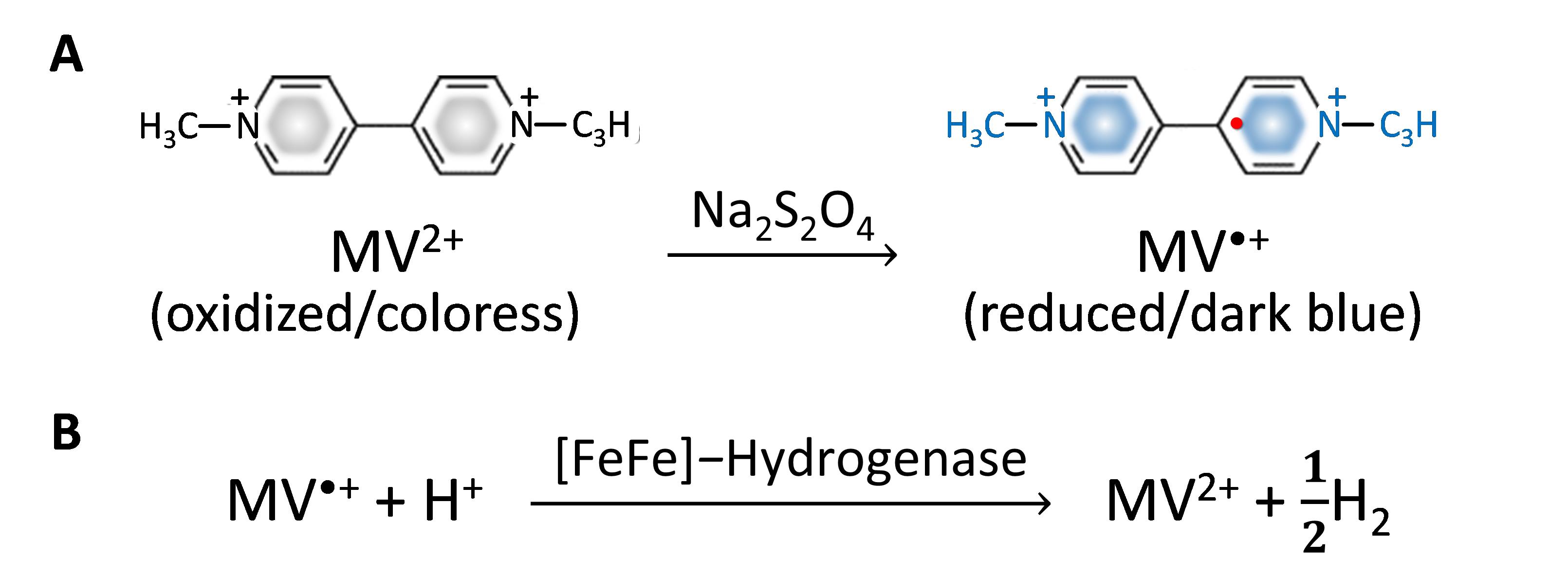
Figure 1. Methyl viologen (MV2+) as an artificial reductant in hydrogenase activity assay. (A) MV2+ reduction to MV•+ formation in Na2S2O4 solution. (B) H2 production from the MV•+-dependent reaction of [FeFe]-hydrogenase.
The amount of H2 generated is directly monitored by gas chromatography. To avoid the detrimental effect of atmospheric oxygen and enhance biocatalytic activity, the reactions are carried out under anaerobic conditions at 37 °C. This method can be applied to determine H2 production capacity of various biological samples such as whole cells, crude cell extracts, or purified enzymes.
Materials and reagents
Biological materials
Hydrogenase-expressing Escherichia coli carrying hydA, hydE, hydF, and hydG genes (this recombinant bacterial strain was constructed in our laboratory according to our previous report in Kosem et al., 2023).
Note: The function of each protein expressed from specific genes in the processes of hydrogenase maturation was reported in many published articles, such as Broderick et al. (2014) and Lubitz et al. (2014).
Reagents
LB broth, Miller (Nacalai Tesque, catalog number: 20068-75)
Methyl viologen (Tokyo Chemical Industry, catalog number: D3685)
Sodium dithionite (Na2S2O4) (Sigma-Aldrich, catalog number: 71699-250G)
PierceTM BCA Protein Assay kit (Thermo Scientific, catalog number: 23227)
Sodium chloride (NaCl) (Wako Chemical, catalog number: 191-01665)
Tris (hydroxymethyl) aminomethane (Tris) (Nacalai Tesque, catalog number: 35434-21)
Hydrochloric acid (HCl) (Wako Chemical, catalog number: 080-0106)
Trichloroacetic acid (TCA) (Nacalai Tesque, catalog number: 34637-14)
3-(N-morpholino)propanesulfonic acid (MOPS) (Nacalai Tesque, catalog number: 23415-25)
Sodium hydroxide (NaOH) (Chameleon Reagent, catalog number: 000-75165)
Ampicillin sodium (Wako, catalog number: 014-23302)
Streptomycin sulfate (Wako, catalog number: 194-08512)
Glucose (Nacalai Tesque, catalog number: 16805-35)
Ferric ammonium citrate (Nacalai Tesque, catalog number: 19425-12)
L-cysteine (Nacalai Tesque, catalog number: 10309-12)
Sodium fumarate (TCI, catalog number: F0070)
Isopropyl-β-D-thiogalactopyranoside (IPTG) (FujiFilm, catalog number: 094-05144)
Solutions
LB (Luria-Bertani) broth (see Recipes)
1 M MOPS-NaOH (see Recipes)
100 mg/mL Ampicillin sodium (see Recipes)
40 mg/mL Streptomycin sulfate (see Recipes)
50% (w/v) Glucose (see Recipes)
250 mg/mL Ferric ammonium citrate (see Recipes)
1 M Sodium fumarate (see Recipes)
1 M IPTG (see Recipes)
0.9% (w/v) NaCl (see Recipes)
50 mM MV2+/Na2S2O4 solution (see Recipes)
1 M Tris-HCl pH 7 (see Recipes)
Recipes
LB broth
Note: Dissolve the medium and all reagents in a 500 mL Erlenmeyer flask and then autoclave at 121 °C for 15 min before use.
Reagent Final concentration Quantity LB broth 2.5 % (w/v) 5 g 1 M MOPS-NaOH (pH 7.4) (Recipe 2) 100 mM 20 mL Water n/a 180 mL Total n/a 200 mL The following (see Recipes 3–8) are added after autoclaving: 100 mg/mL Ampicillin sodium 100 μg/mL 0.2 mL 40 mg/mL Streptomycin sulfate 40 μg/mL 0.2 mL 50% (w/v) Glucose 0.5% (w/v) 2 mL 250 mg/mL Ferric ammonium citrate 250 μg/mL 0.2 mL L-cysteine 2 mM 50 mg 1 M Sodium fumarate 20 mM 4 mL 1 M IPTG 1 mM 0.2 mL 1 M MOPS-NaOH pH 7.4
Note: Dissolve in a 100 mL beaker on a magnetic stirrer.
Reagent Final concentration Quantity MOPS 1 M 20.9 g Water n/a 80 mL NaOH (8 M) n/a Slowly add until pH 7.4 Total n/a Make up final volume to 100 mL 100 mg/mL Ampicillin sodium
Note: Dissolve in a 5 mL microcentrifuge tube and mix thoroughly with gentle shaking by hand. Aliquot the stock solution in 1 mL microcentrifuge tubes (500 μL per tube) and store at -20 °C until use.
Reagent Final concentration Quantity Ampicillin sodium 100 mg/mL 0.5 g Sterilized water n/a 5 mL Total n/a 5 mL 40 mg/mL Streptomycin sulfate
Note: Dissolve in a 5 mL microcentrifuge tube and mix thoroughly with gentle shaking by hand. Aliquot the stock solution in 1 mL microcentrifuge tubes (500 μL per tube) and store at -20 °C until use.
Reagent Final concentration Quantity Streptomycin sulfate 40 mg/mL 0.2 g Sterilized water n/a 5 mL Total n/a 5 mL 50% (w/v) Glucose
Note: Dissolve glucose powder with warm sterilized water in a 100 mL beaker on a magnetic stirrer. Once it has completely dissolved, bring the volume up to 100 mL total. Aliquot the stock solution in 15 mL tubes (10 mL per tube) and store at -20 °C until use.
Reagent Final concentration Quantity Glucose 50% (w/v) 50 g Sterilized water n/a 60 mL Total n/a Make up final volume to 100 mL 250 mg/mL Ferric ammonium citrate
Note: Dissolve in a 5 mL microcentrifuge tube and mix thoroughly with gentle shaking by hand. Aliquot the stock solution in 1 mL microcentrifuge tubes (500 μL per tube) and store at -20 °C until use.
Reagent Final concentration Quantity Ferric ammonium citrate 250 mg/mL 1.25 g Sterilized water n/a 5 mL Total n/a 5 mL 1 M Sodium fumarate
Note: Dissolve in a 50 mL tube and mix thoroughly with gentle shaking by hand. Aliquot the stock solution in 15 mL tubes (10 mL per tube) and store at -20 °C until use.
Reagent Final concentration Quantity Sodium fumarate 1 M 8.0 g Sterilized water n/a 50 mL Total n/a 50 mL 1 M IPTG
Note: Dissolve in a 5 mL microcentrifuge tube and mix thoroughly with gentle shaking by hand. Aliquot the stock solution in 1 mL microcentrifuge tubes (500 μL per tube) and store at -20 °C until use.
Reagent Final concentration Quantity IPTG 1 M 1.2 g Sterilized water n/a 5 mL Total n/a 5 mL 0.9% (w/v) NaCl solution
Note: Dissolve in a 100 mL beaker on a magnetic stirrer.
Reagent Final concentration Quantity NaCl 0.9% (w/v) 0.9 g Water n/a 100 mL Total n/a 100 mL 50 mM MV2+/Na2S2O4 solution
Note: Prepare fresh in an anaerobic sealed vial inside a glove box and mix thoroughly with gentle shaking by hand.
Reagent Final concentration Quantity Methyl viologen 50 mM 0.0257 g Na2S2O4 250 mM 0.0870 g Water n/a 2 mL Total n/a 2 mL 1 M Tris-HCl pH 7
Note: Dissolve in a 100 mL beaker on a magnetic stirrer. Adjust pH with 3 M HCl in a fume hood and strictly following the Kyushu University Guidelines for Safety in Education: Laboratory Activities. A safety data sheet from the chemical company as shown in Guideline 1 in the Supporting materials.
Reagent Final concentration Quantity Tris 1 M 12.1 g Water n/a 80 mL HCl (3 M) n/a Slowly add until pH 7 Total n/a Make up final volume to 100 mL
Laboratory supplies
Glass vial No. 3, 10 mL size (ASONE, catalog number: 5-111-03)
Rubber stopper (ASONE, catalog number: 5-112-01)
Aluminum cap (ASONE, catalog number: 5-112-03)
1 mL Syringe (Terumo, catalog number: SS-01T)
Needle No. 27 G × 3/4" (Terumo, catalog number: NN-2719S)
10 μL pipette tip (Molecular BioProducts, catalog number: 3510-05)
200 μL pipette tip (Violamo, catalog number: 3-6504-12)
1,000 μL pipette tip (Violamo, catalog number: 3-6504-13)
1 mL microcentrifuge tube (Quality Scientific Plastics, catalog number: L-510-GRD-Q)
5 mL microcentrifuge tube (ASONE, model: AST0500)
96-well plate (ASONE, catalog number: 2-8085-02)
99.999% N2 gas (Air Liquide, catalog number: JAGA 13038)
Gloves (Showaglove, catalog number: 882.L.BLUE)
Paper towels
Disposable cuvettes (Violamo, model: UVC-Z8.5)
Equipment
2–20 μL autoclavable micropipette (Nichipet Ex II, Nichiryo, catalog number: J16Y01961)
20–200 μL autoclavable micropipette (Nichipet Ex II, Nichiryo, catalog number: J16Z00871)
100–1,000 μL autoclavable micropipette (Nichipet Ex II, Nichiryo, catalog number: J16X11751)
accu-jet® pro pipette controller (BRAND®, catalog number: Z671533)
High-speed micro centrifuge (Hitachi, model: CF16RN)
Personal centrifuge (Front Lab, model: FLD2012)
Gas chromatography (Shimadzu Corp., model: GC-8A)
Thermal conductivity detector (Shimadzu Corp., model: GC-8AIT)
Integrator C-R6A chromatopac (Shimadzu Corp., catalog number: 223-04500-38)
Molecular sieve 5A beads (GL Sciences Inc., catalog number: 1001-11503)
Stainless steel column, 2 m length × 3 mm diameter (Shimadzu Corp., catalog number: 201-48705-20)
Shaking water bath (Yamato Scientific, model: BW101)
Shaking incubator (EYELA, model: FYC-100)
Anaerobic glove box (MIWA, model: 1ADB-3)
Clean bench (Hitachi Appliances Inc., model: CCV-1300E)
Microplate reader (Corona Electric, model: SH-1000)
500 mL Erlenmeyer flask with baffle (IWAKI, model: CTE33)
250 mL centrifuge bottle (Nalgene, catalog number: B1033)
20 mm vial crimper (Chromatography Research Supplies, catalog number: 320990)
20 mm vial decapper pliers (Kebby, catalog number: D-20)
1 mL gas tight syringe (ITO Corporation, catalog number: MS-GAN100)
Weighing balance with 0.0001 g accuracy (Metter Toledo, model: XS204)
Vortex mixer (Scientific Industries, model: SI-0286)
pH meter (Horiba Scientific, model: 9625)
Water distillation apparatus (Advantec, model: RFD240NA)
Magnetic stirrer (Pasorina Stirrer, model: CT-MINI)
100 mL Beaker (AGC Iwaki, model: CTE33)
Fume hood (Oriental, model: TNV-STZ-1800HCS)
Software and datasets
Microsoft Excel
SF6 (version 5.6.0, Copyright© 2005 Corona Electric) for spectrophotometer
Procedure
Cultivation of hydrogenase-expressing bacteria
In this protocol, a recombinant E. coli encoding hydA, hydE, hydF, and hydG genes for hydrogenase expression was utilized as a H2-producing model constructed in our laboratory as reported in Honda et al. (2016) and Kosem et al. (2023).
Note: This protocol can be applied to determine the efficiency of other hydrogenase-producing bacteria, as reported in Benoit et al. (2020) and Kosem et al. (2024).
Aerobically pre-culture the recombinant E. coli in a 500 mL Erlenmeyer flask with 200 mL of LB broth supplemented with 100 μg/mL of ampicillin sodium, 40 μg/mL of streptomycin sulfate, 0.5% (w/v) of glucose, 250 μg/mL of ferric ammonium citrate, and 100 mM MOPS/NaOH pH 7.4. Incubate the pre-culture flask at 37 °C placed on a shaking incubator at 120 rpm until the cell density reaches an optical density of 0.4 at OD600, monitored by a spectrophotometer using a 1 mL cell suspension in a disposable cuvette. Then, transfer the 200 mL pre-cultured E. coli suspension into a 250 mL centrifuge bottle inside an anaerobic glove box supplemented with 2 mM L-cysteine (50 mg), 20 mM sodium fumarate (4 mL of 1 M stock solution), and 1 mM IPTG (0.2 mL of 1 M stock solution), and further incubate for 18 h in the glove box for [FeFe]-hydrogenase expression (Figure 2A).
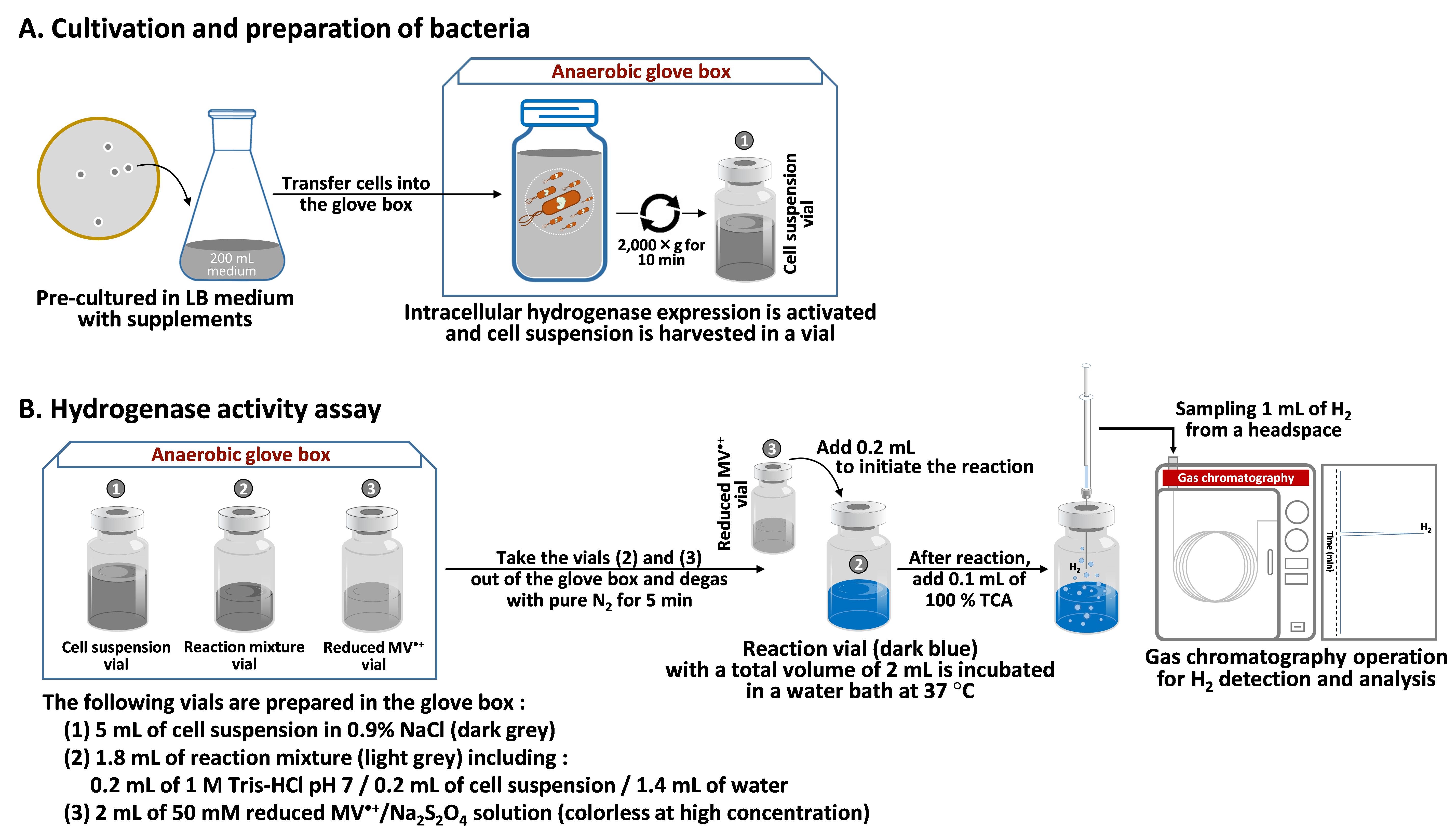
Figure 2. Experimental process of MV2+-dependent hydrogenase activity
Cell harvesting and preparation
In the glove box, transfer 200 mL of cell suspension cultivated in LB medium from the Erlenmeyer flask to a centrifuge bottle with a silicone cap to preserve an anaerobic environment.
Centrifuge the bottle at 2,000× g for 10 min, discard the supernatant, and collect cell pellet.
Wash the cell pellet once with 5 mL of 0.9% NaCl solution and centrifuge at 2,000× g for 10 min.
To prepare cell suspension for experiments, resuspend the washed cell pellet in 5 mL of 0.9% NaCl solution (Figure 2A).
To standardize the protocol, the concentration of cells used in the reaction is based on total protein contents measured by PierceTM BCA Protein Assay kit in a 96-well plate.
Note: The protocol can be modified when different bacterial strains or biological samples are used.
Hydrogenase activity assay
Degas all chemical reagents with pure N2 for 5 min and place them in the anaerobic glove box before use.
Prepare the following reagents as shown in Table 1 and Figure 2B.
Table 1. Reaction mixture preparation of MV•+-hydrogenase activity assay
Reagents Applied volume Final concentration 1 M Tris-HCl pH 7 0.2 mL 100 mM 10 mg/mL protein of cell suspension or extract 0.2 mL 1 mg/mL Sterile deionized water 1.4 mL 50 mM MV2+ in 250 mM Na2S2O4 (reduced MV•+ solution) 0.2 mL 5 mM Total 2 mL
Notes:The reaction mixture is prepared in a 10 mL glass vial and sealed with a rubber stopper and aluminum cap inside the glove box.
The protein concentration can be adjusted in different samples.
In case of a negative control, the same volume of sterile water is added instead of cell suspension or extracts.
The reduced MV•+ solution is prepared in a separate vial and sealed with a rubber stopper and aluminum cap inside the glove box.
Take the reaction vial and the MV•+ vial out of the glove box.
Purge the contaminated gases in each vial with pure N2 for 5 min.
Initiate the reaction by adding 0.2 mL of 50 mM MV2+/Na2S2O4 solution into the reaction vial using a 1 mL syringe with needle No. 27 G × 3/4" and further incubate in a shaking water bath at 100 rpm and 37 °C.
Note: The reaction mixture turns dark blue after adding MV2+/Na2S2O4 solution into the reaction vial.
Every 20 min after incubation, terminate the reaction vial by adding 0.1 mL of 100% TCA (concentration of the commercial stock solution) using a 1 mL syringe with needle No. 27 G × 3/4".
Sample 1 mL of H2 produced in a headspace of the reaction vial using a 1 mL gas tight syringe and vertically inject into a gas chromatograph for analysis. Here, a GC-8A gas chromatograph equipped with a thermal conductive detector and an integrator C-R6A chromatopac was used. The produced gas goes through molecular sieve 5A beads packed in a stainless steel column (2 m length × 3 mm diameter) with a carrier gas of argon. Operating parameters are shown in Table 2.
Table 2. Chromatographic operating parameters
Parameters Units Pressure of carrier gas, Ar 100 kPa Injection volume 1 mL Detector temperature 50 °C Injection temperature 50 °C Column temperature 50 °C Detector current 60 mA
Data analysis
Quantification of H2:
The accurate amount of H2 produced from the reaction is calculated from a calibration curve between H2 concentration vs. peak area obtained from the chromatogram of GC analysis (Figure 3).
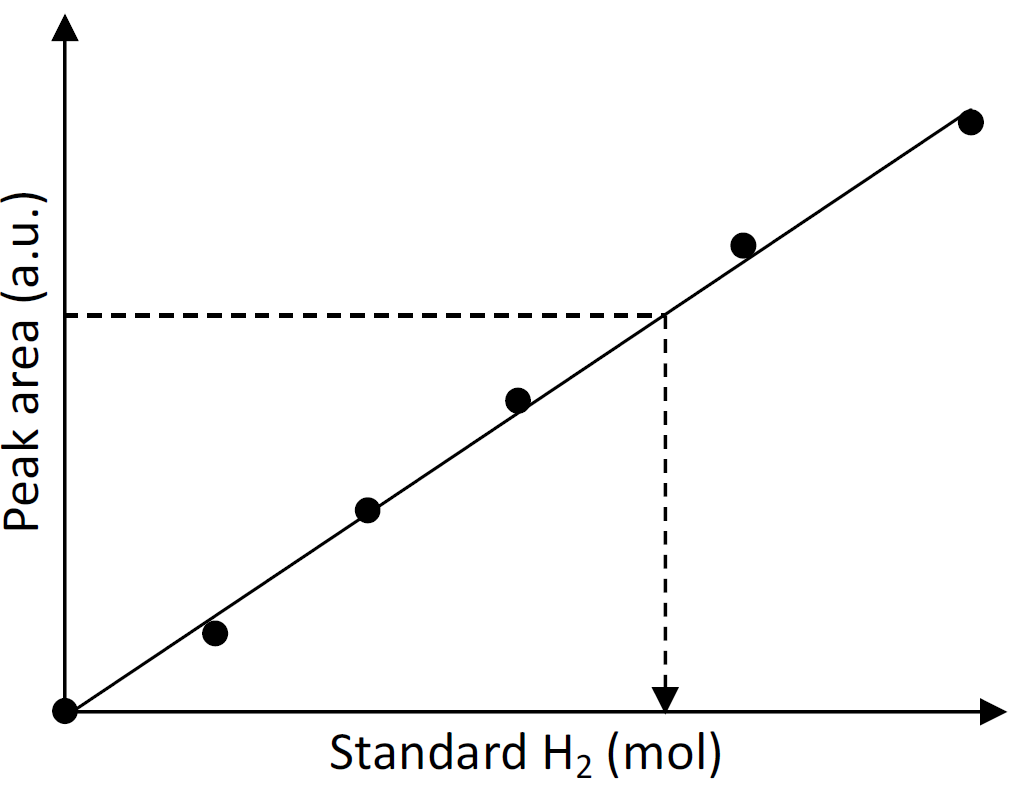
Figure 3. Calibration plot of standard H2 vs. peak area by gas chromatography. According to the calibration curve, the amount of H2 can be calculated from the following equation: H2 (mol) = (Peak area – Intercept)/Slope.Calculation of hydrogenase activity
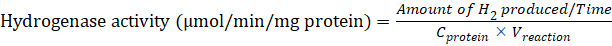 (1)
(1)In which the amount of H2 (μmol) in the headspace (Figure 4A) is measured at each time point and plotted to obtain the H2 production rate (Figure 4B).
Time (minutes or hours) is the incubation time of the reaction.
Note: Normally it takes 1–2 h for the incubation period. The amount of H2 production rate is presented in μmol per min.
Cprotein (mg/mL) is the final concentration of protein in the reaction mixture.
Vreaction (mL) is the total volume of the reaction mixture.
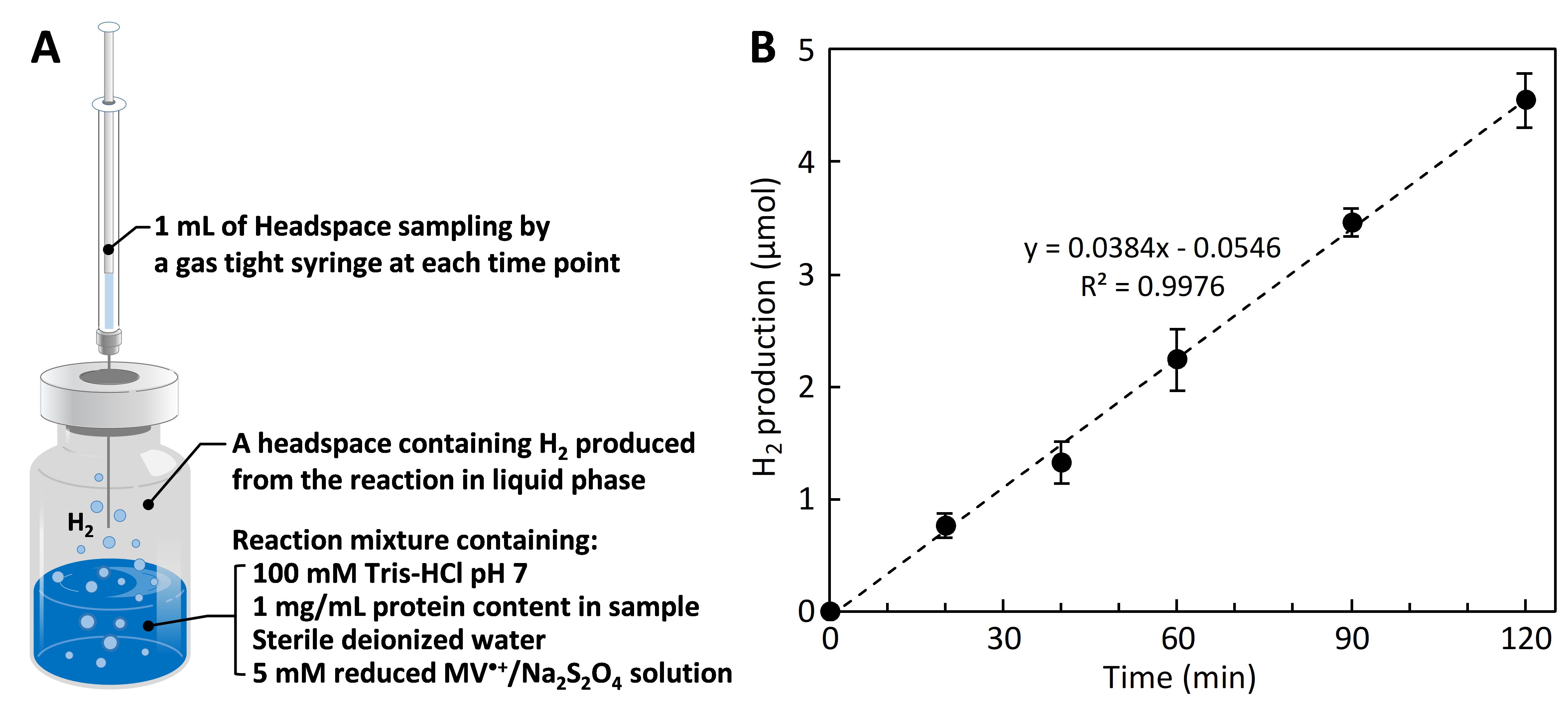
Figure 4. Methyl viologen (MV•+)-dependent hydrogenase activity assay. (A) Reaction vial containing the liquid phase of reaction mixture and the gas phase of H2 produced in a headspace. (B) H2 production rate is calculated from a kinetic plot between the amount of H2 in the y-axis vs. time in the x-axis. Hydrogenase activity is calculated from equation (1).
Validation of protocol
This protocol was validated in Kosem et al. (2023). Applied Catalysis A: General, DOI: 10.1016/j.apcata.2022.119019.
General notes and troubleshooting
To preserve the biological function of O2-sensitive enzymes, biological samples containing hydrogenase must be protected from air. Therefore, the whole process of cell preparation and experiments must be performed under anaerobic environment or in the glovebox.
Kinetic parameters of different hydrogenases from various biological sources can be analyzed by varying the concentrations of methyl viologen as a substrate according to the Michaelis-Menten equation:
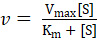 , where v is velocity, Vmax is the maximum velocity, Km is the Michaelis constant, and [S] is the concentration of the substrate (methyl viologen).
, where v is velocity, Vmax is the maximum velocity, Km is the Michaelis constant, and [S] is the concentration of the substrate (methyl viologen).
Acknowledgments
I would like to thank Prof. Tatsumi Ishihara for his supervision on my previous research, which originally validated this protocol in Kosem et al. (2023) that was funded by a Grant-in-Aid for Specially Promoted Research (No. 21K18213) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan through the Japan Society for the Promotion of Science. I would also like to acknowledge the Department of Applied Chemistry, Faculty of Engineering, Kyushu University, and the International Institute for Carbon-Neutral Energy Research (I2CNER), Kyushu University for research facilities and instruments. I am thankful to Dr. Yuki Honda from whom I adapted this protocol.
Competing interests
The authors declare no competing interests.
References
Benoit, S. L., Maier, R. J., Sawers, R. G. and Greening, C. (2020). Molecular Hydrogen Metabolism: a Widespread Trait of Pathogenic Bacteria and Protists. Microbiol. Mol. Biol. Rev. 84(1): e00092-19.
- Broderick, J. B., Byer, A. S., Duschene, K. S., Duffus, B. R., Betz, J. N., Shepard, E. M. and Peters, J. W. (2014). H-Cluster assembly during maturation of the [FeFe]-hydrogenase. J. Biol. Inorg. Chem. 19(6): 747–757.
- Honda, Y., Hagiwara, H., Ida, S. and Ishihara, T. (2016). Application to Photocatalytic H2 Production of a Whole‐Cell Reaction by Recombinant Escherichia coli Cells Expressing [FeFe]‐Hydrogenase and Maturases Genes. Angew. Chem. Int. Ed. 55(28): 8045–8048.
- Kosem, N., Watanabe, M., Song, J. T., Takagaki, A. and Ishihara, T. (2023). A comprehensive study on rational biocatalysts and individual components of photobiocatalytic H2 production systems. Appl. Catal. A: Gen. 651: 119019.
- Kosem, N., Shen, X.-F., Ohsaki, Y., Watanabe, M., Song, J. T. and Ishihara, T. (2024). Photobiocatalytic conversion of solar energy to NH3 from N2 and H2O under ambient condition. Appl. Catal. B: Environ. 342: 123431.
- Lubitz, W., Ogata, H., Rüdiger, O. and Reijerse, E. (2014). Hydrogenases. Chem. Rev. 114(8): 4081–4148.
- Ogo, S., Kishima, T., Yatabe, T., Miyazawa, K., Yamasaki, R., Matsumoto, T., Ando, T., Kikkawa, M., Isegawa, M., Yoon, K. S., et al. (2020). [NiFe], [FeFe], and [Fe] hydrogenase models from isomers. Sci. Adv. 6(24): eaaz8181.
- Orgill, J. J., Chen, C., Schirmer, C. R., Anderson, J. L. and Lewis, R. S. (2015). Prediction of methyl viologen redox states for biological applications. Biochem. Eng. J. 94: 15–21.
- Tard, C. and Pickett, C. J. (2009). Structural and Functional Analogues of the Active Sites of the [Fe]-, [NiFe]-, and [FeFe]-Hydrogenases. Chem. Rev. 109(6): 2245–2274.
- Xuan, J., He, L., Wen, W. and Feng, Y. (2023). Hydrogenase and Nitrogenase: Key Catalysts in Biohydrogen Production. Molecules 28(3): 1392.
Supplementary information
The following supporting information can be downloaded here:
- Figure S1. Catalytic sites of different hydrogenases.
- Guideline 1. Safety procedure in handling HCl.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Kosem, N. (2023). H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography. Bio-protocol 13(23): e4895. DOI: 10.21769/BioProtoc.4895.
Category
Biochemistry > Protein > Activity
Microbiology > Microbial biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










