- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Phospholipid Preparations to Characterize Protein–Lipid Interactions In Vitro
Published: Vol 13, Iss 22, Nov 20, 2023 DOI: 10.21769/BioProtoc.4887 Views: 1796
Reviewed by: Julie WeidnerJohn P PhelanPhilipp A.M. Schmidpeter

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Enrichment of Membrane Proteins for Downstream Analysis Using Styrene Maleic Acid Lipid Particles (SMALPs) Extraction
Benedict Dirnberger [...] Kathryn S. Lilley
Aug 5, 2023 2978 Views

Real-Time Monitoring of ATG8 Lipidation in vitro Using Fluorescence Spectroscopy
Wenxin Zhang [...] Sharon A. Tooze
Jan 5, 2024 1872 Views

Lipid-Mediated Sequential Recruitment of Proteins Via Dual SLIPT and Dual SLIPTNVOC in Live Cells
Kristina V. Bayer and Richard Wombacher
Nov 5, 2025 1603 Views
Abstract
The lipid bilayers of the cell are composed of various lipid classes and species. These engage in cell signaling and regulation by recruiting cytosolic proteins to the membrane and interacting with membrane-embedded proteins to alternate their activity and stability. Like lipids, membrane proteins are amphipathic and are stabilized by the hydrophobic forces of the lipid bilayer. Membrane protein–lipid interactions are difficult to investigate since membrane proteins need to be reconstituted in a lipid-mimicking environment. A common and well-established approach is the detergent-based solubilization of the membrane proteins in detergent micelles. Nowadays, nanodiscs and liposomes are used to mimic the lipid bilayer and enable the work with membrane proteins in a more natural environment. However, these protocols need optimization and are labor intensive. The present protocol describes straightforward instructions on how the preparation of lipids is performed and how the lipid detergent mixture is integrated with the membrane protein MARCH5. The lipidation protocol was performed prior to an activity assay specific to membrane-bound E3 ubiquitin ligases and a stability assay that could be used for any membrane protein of choice.
Keywords: Membrane protein–lipid interactionBackground
Membrane proteins interact with their lipid environment with the hydrophobic and hydrophilic patches of their transmembrane domains via a specific binding site (non-annular lipids) or the physiochemical properties of the lipid bilayer (annular lipids) (Contreras et al., 2011; Stangl and Schneider, 2015). These protein–lipid interactions can alter the activity, stability, folding, or localization of the target protein, leading to various cellular signaling events (Engelman, 2005; van Meer et al., 2008; Corradi et al., 2019a;). The characterization of these signaling mechanisms and the corresponding lipid–membrane protein interaction is a constant challenge due to the inherent phase difference imposed on the system from the presence of a mixture of detergent/lipid, lipid/protein, and detergent/lipid/protein (Sych et al., 2022). Therefore, research into the mechanisms used by lipids to regulate protein function and structure is restricted to a small number of membrane protein systems, often because of the technical difficulties as well as complexity associated with handling these systems in a laboratory setting (Fahy et al., 2009). Hence, understanding the mechanism of how lipids can modulate the activity of a specific membrane protein and the identification of the lipids that interact with the membrane protein of interest is often still an open and fundamental research question (Corradi et al., 2019b).
Phospholipids are the most abundant lipids in mammalian membranes and are characterized by their hydrophilic head group and their hydrophobic tail, which is built by two fatty acid chains. These fatty acid chains can vary in length and saturation, creating an immense variety (Horvath and Daum, 2013; Cockcroft, 2021). The chemically diverse hydrophilic head groups define the lipid classes (Table 1), whereas the different lengths and saturation subdivide the class in different lipid species (Liebisch et al., 2020). The lipid composition of the lipid bilayer is specific to the tissue and the cellular compartment, which comprises a distinct set of phospholipids (Table 1) (Ernst et al., 2016).
Table 1. Overview of phospholipids, their chemical characteristic, and the abundance of subcellular fractions [plasma membrane, mitochondria, and endoplasmic reticulum (ER)) of rat liver (Horvath and Daum, 2013)]
| Phospholipid class | Characteristic | Abundance in cellular compartment (%) | ||
|---|---|---|---|---|
| Plasma membrane | Mitochondria | ER | ||
| Phosphatidylethanolamine (PE) | zwitterionic | 40 | 34 | 60 |
| Cardiolipin (CL) | anionic | 1 | 14 | 1 |
| Phosphatidic acid (PA) | anionic | 1 | < 1 | 1 |
| Phosphatidylcholine (PC) | zwitterionic | 24 | 44 | 23 |
| Phosphatidylglycerol (PG) | anionic | - | - | - |
| Phosphatidylinositol phosphate (PI) | anionic | 8 | 5 | 10 |
Nowadays, different membrane-mimicking systems are available to solvate membrane proteins. The most common method for working with membrane proteins in vitro is to solubilize them in detergent. The detergent forms a micelle layer around the hydrophobic surface patches of the protein, leading to the stabilization of the membrane (or membrane-associated) protein outside the natural lipid bilayer. Over the last years, alternatives to detergent-based methods were developed, such as nanodiscs (Pettersen et al., 2023), styrene–maleic acid copolymers (SMAs) (Dörr et al., 2016), or liposomes (Verchère et al., 2017). However, these methods often need a detergent-solubilized membrane protein as the initial step and optimization is often labor intensive. Here, we describe two easy-to-establish and straightforward protocols to examine the interaction and effect of different lipid classes on the activity and stability of purified and detergent-solubilized MARCH5 in vitro. The described protocols were initially used to test lipid stability in connection with the human integral membrane protein SERINC5 (Pye et al., 2020) and, since then, tested against a broader spectrum of membrane proteins (Cecchetti et al., 2021).
Materials and reagents
Biological materials
Protein of interest (a membrane protein solubilized in detergent); in this protocol, using MARCH5 as benchmarking example (see Merklinger et al., 2022).
Reagents
Lipid preparation for activity and stability assays
Egg PE (Avanti Polar Lipids, catalog number: 840021)
Heart CL (Avanti Polar Lipids, catalog number: 840012)
Egg PA (Avanti Polar Lipids, catalog number: 840101)
Egg PC (Avanti Polar Lipids, catalog number: 840051)
Egg PG (Avanti Polar Lipids, catalog number: 841138)
Brain PI(4)P (Avanti Polar Lipids, catalog number: 840045)
Lauryl maltose neopentyl glycol (LMNG) (Anatrace, catalog number: NG310)
Argon gas
Lipid-binding assay
LMNG (Anatrace, catalog number: NG310)
Trizma base (Sigma-Aldrich, catalog number: 77-86-1)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653)
BSA (Sigma-Aldrich, catalog number: A9418)
PierceTM ECL western blotting substrate (Thermo Fischer Scientific, catalog number 32132)
Amersham HydondTM-LFP, Hybond LFP polyvinylidene fluoride or polyvinylidene difluoride (PVDF) transfer membrane (GE Healthcare, catalog number: RPN303LFP)
Egg PE (Avanti Polar Lipids, catalog number: 840021)
Heart CL (Avanti Polar Lipids, catalog number: 840012)
Egg PA (Avanti Polar Lipids, catalog number: 840101)
Egg PC (Avanti Polar Lipids catalog number: 840051)
Egg PG (Avanti Polar Lipids catalog number: 841138)
Brain Phosphatidylinositol-4-phosphate (PI(4)P) (Avanti Polar Lipids, catalog number: 840045)
Anti-MARCH5 antibody N-terminal (Abcam, catalog number: ab185054)
Anti-rabbit IgG, HRP-linked Antibody (Cell Signalling, catalog number: 7074)
Solutions
0.25% LMNG (see Recipes)
Tris-buffered saline (see Recipes)
Tris-buffered saline supplemented with 0.001% LMNG (see Recipes)
Blocking solution (TBS supplemented with 0.001% LMNG and 3% BSA) (see Recipes)
500× diluted Anti-MARCH5 antibody in TBS supplemented with 0.001% LMNG and 3% BSA (see Recipes)
2,000× diluted Anti-rabbit IgG antibody in TBS supplemented with 0.001% LMNG and 3% BSA (see Recipes)
Recipes
0.25% LMNG
Reagent Final concentration Quantity LMNG 0.25% (w/v) 0.0025 g H2O n/a 10 mL Total 0.25% 10 mL TBS (Tris-buffered saline)
Reagent Final concentration Quantity Trizma base 50 mM 6.05 g NaCl 150 mM 8.76 g H2O 1,000 mL Total 1,000 mL pH is adjusted to 7.5 using HCl TBS supplemented with 0.001% LMNG
Reagent Final concentration Quantity TBS (see Recipe 2) 30 mL 0.25% LMNG (see Recipe 1) 0.001% 0.12 mL Total 30 mL Blocking solution
Reagent Final concentration Quantity TBS (see Recipe 2) 30 mL 0.25% LMNG (see Recipe 1) 0.001% 0.12 mL BSA 3% 0.9 g Total 30 mL 500× diluted Anti-MARCH5 in TBS supplemented with 0.001% LMNG and 3% BSA
Reagent Final concentration Quantity Blocking solution 5 mL Anti-MARCH5 antibody 1:500 dilution 0.01 mL Total 5 mL 2,000× diluted Anti-rabbit IgG antibody in TBS supplemented with 0.001% LMNG and 3% BSA
Reagent Final concentration Quantity Blocking solution 5 mL Anti-rabbit IgG antibody 1:2,000 dilution 0.0025 mL Total 5 mL
Laboratory supplies
FisherbrandTM Borosilicate glass light-walled culture tubes (Fisher Scientific, catalog number: 11517403, diameter 12 mm, length 75 mm)
Eppendorf tubes (Eppendorf, catalog number: 0030120086)
Equipment
Pulsing Vortex mixer (VWR, catalog number: 10153-812)
Plate shaker (VWR, catalog number: 700-0246)
ChemiDocTM Touch Gel imaging system (Bio-Rad, catalog number: 1708370)
Procedure
Lipid preparation
Fill round-bottom glass tubes with argon gas.
Transfer 20 μL of Egg PE, Heart CL, Egg PA, Egg PG, and Egg PC chloroform solutions and 50 μL of Brain PI(4)P chloroform solution (all stored at -20 °C under argon gas) to a separate argon gas–filled round-bottom glass tube. If lipid combinations are to be tested, it is possible to dry the lipids in the same round-bottom tube in the desired amounts. This reduces the detergent concentration in the protein–lipid mix.
Dry the lipids under an argon stream and shake on a pulsing vortex mixer for 15 min.
Re-suspend dry lipids to 5 μM under shaking, depending on their average molecular weight:
Egg PE (MW: 746 Da) in 54 μL of 0.25% LMNG.
Heart CL (MW: 1494 Da) 27 μL of 0.25% LMNG.
Egg PA (MW: 711 Da) 57 μL of 0.25% LMNG.
Egg PC (MW: 770 Da) 52 μL of 0.25% LMNG.
Egg PG (MW: 782) 51 μL of 0.25% LMNG.
Brain PI(4)P (MW: 974) 10 μL of 0.25% LMNG.
Add 1.8 μL of the 5 μM lipid solution to 50 μL of 32 μM purified MARCH5 (1 mg/mL) in 100 mM NaCl; 50 mM Tris pH 7.7; 5% glycerol; 0.001% LMNG; 0.1 mM TCEP (MARCH5 is incubated with the different lipids in a 1:10 molar ratio).
Incubate the lipid–protein solution on ice for 1 h.
The protein is now considered ready for activity and/or stability measurements.
Important: work in a laminar flow hood when working with chloroform and argon gas.
Note: Generate lipid stocks that take the lipid:detergent ratio into consideration in the final assay conditions. Thus, the lipid stocks might have to be adjusted to keep the reaction mixture volumes equal, while still keeping the final critical micelle concentration (CMC) equivalent throughout the assay. This can be done by adjusting the detergent:lipid ratios already in the stock solution. However, this also limits the maximal detergent:lipid ratio that can be tested in solution.
Lipid-binding assay (Figure 1A)
Cut a rectangle (2 cm × 6 cm) of the PVDF membrane.
Spot 1 μL of the chloroform solutions of Egg PE, Heart CL, Egg PA, Egg PC, Egg PG, and Brain PI(4)P on the PVDF membrane at approximately 1 cm of distance.
Dry the spotted lipid for 1 h at room temperature (RT) in a laminar flow hood.
Spot positive controls on the dry membrane lipid strip:
Spot 1 μL of Anti-rabbit IgG, HRP-linked antibody on the membrane lipid strip.
Spot 1 μL of 0.35 mg/mL target protein.
Dry positive controls for 15 min.
Block the PVDF membrane with 30 mL of TBS supplemented with 3% BSA overnight at 4 °C with soft agitation on a plate shaker.
Remove the blocking solution.
Add 10 mL of 1 μg/mL of MARCH5 in TBS supplemented with 0.001% LMNG and 3% BSA (1 CMC of LMNG is added to ensure MARCH5 will not precipitate during incubation) to the PVDF membrane.
Incubate the PVDF membrane and MARCH5 for 4 h at 4 °C with soft agitation on a plate shaker.
Wash the PVDF membrane three times with 5 mL of TBS supplemented with 0.001% LMNG for 5 min at RT under soft agitation.
Add 5 mL of Anti-MARCH5 antibody N-terminal in TBS supplemented with 0.001% LMNG and 3% BSA to the PVDF membrane.
Incubate anti-MARCH5 antibody N-terminal for 4 h at 4 °C.
Remove the antibody solution and rinse the PVDF with 5 mL of TBS supplemented with 0.001% LMNG.
Add anti-rabbit IgG, HRP-linked Antibody in 5 mL of TBS supplemented with 0.001% LMNG and 3% BSA to the PVDF membrane.
Wash again the membrane lipid strip three times in 5 mL of TBS supplemented with 0.001% LMNG for 5 min at RT under soft agitation.
Prepare PierceTM ECL western blotting substrate, add onto the membrane lipid strip, and incubate for 1 min.
Measure chemiluminescence using the ChemiDocTM Touch Gel imaging system (Figure 1B and 1C).
Note: This assay will give you a yes/no answer on lipid membrane protein binding (see Figure 1).
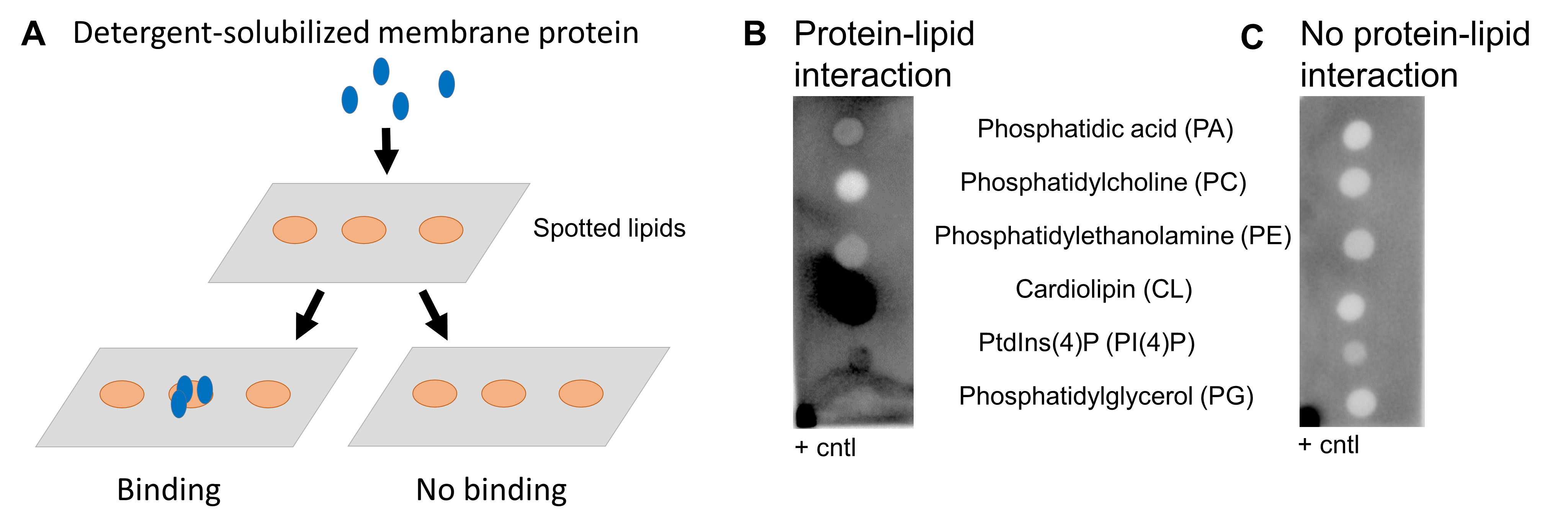
Figure 1. Lipid binding assay. A. Schematic overview of lipid binding assay. B. Example of positive hits. Cardiolipin and PI4P interact with the protein of interest, indicated by the dark dots. The protein of interest spotted in the left-right corner is a positive control (+ cntl). C. Example of no lipid-protein interaction. The positive control (+ cntl) shows a positive signal indicating that the assay has worked properly.
Validation of protocol
Lipid preparation
The phospholipids prepared as described in this protocol were used in differential scanning fluorimetry as well as ubiquitination assays published in the article “Phospholipids alter activity and stability of mitochondrial membrane-bound ubiquitin ligase MARCH5”, Life Science Alliance Apr 2022, DOI: 10.26508/lsa.202101309 (see Figure 1D, Figures 2 and 4) (Merklinger et al., 2022). The experiments were performed in three independent experiments and showed consistent results. Similar experiments were performed by Cecchetti et al., investigating protein stability in different lipids (Cecchetti et al., 2021).
Lipid-binding assay
The lipid-binding assay described in this protocol was used to investigate the binding of MARCH5 to various lipids published in the article “Phospholipids alter activity and stability of mitochondrial membrane-bound ubiquitin ligase MARCH5”, Life Science Alliance Apr 2022, DOI: 10.26508/lsa.202101309 (see Figure 1B and S1). It was also used to investigate DRP1 interaction with PA (Adachi et al., 2016). Furthermore, commercially available lipid membrane strips are available from Echelon Biosciences Inc. (https://www.echelon-inc.com/product/membrane-lipid-strips).
General notes and troubleshooting
General notes
Lipid preparation
The purchased phospholipid classes are a mixture of different lipid species, meaning they vary in chain length and saturation (e.g., CL is a mix of 18:1 and 18:2 fatty acid chains) and are from natural origin. This allows us to study the whole lipid class and not only a specific lipid species. However, specific lipid species can be purchased as well. Purchasing lipids dissolved in chloroform makes the handling of the lipids easier (to avoid weighing out small quantities).
Changes in experimental output over time could be due to oxidized phospholipids. It is important to store the phospholipids under inert conditions at -20 °C (under argon or nitrogen).
Lipid binding assay
During the incubation and wash steps, the membrane should always be kept wet and never dry out.
Mark the membrane by cutting one edge to be aware of the orientation and the order of the spotted lipids.
Troubleshooting
Lipid binding assay
The best protein concentration for the lipid-binding assay has to be experimentally determined. A good starting concentration is 1 mg/mL.
The incubation time will need to be optimized for each protein of choice. If the incubation of the membrane protein is kept for too long, this might affect the stability of the protein, and this could lead to errors in the stability measurement and loss of reproducibility. At high membrane protein concentrations, aggregational effect might influence the thermal shift assay measurement; this could also lead to loss of reproducibility.
It is advisable to test a subset of different buffer conditions, i.e., testing the high and low concentrations of the desired salt in combination with variable pH ranges, to ensure that the average state of the membrane protein remain consistent during the time in which the assay is performed.
Acknowledgments
The protocol was first published in Life Science Alliance in April 2022 (DOI: 10.26508/lsa.202101309). We thank all authors for their contribution to this work and the Technical University of Denmark for supporting the project.
Competing interests
The authors declare that they have no conflicts of interest.
References
Adachi, Y., Itoh, K., Yamada, T., Cerveny, K. L., Suzuki, T. L., Macdonald, P., Frohman, M. A., Ramachandran, R., Iijima, M., Sesaki, H., et al. (2016). Coincident Phosphatidic Acid Interaction Restrains Drp1 in Mitochondrial Division. Mol. Cell 63(6): 1034–1043.
- Cecchetti, C., Strauss, J., Stohrer, C., Naylor, C., Pryor, E., Hobbs, J., Tanley, S., Goldman, A. and Byrne, B. (2021). A novel high-throughput screen for identifying lipids that stabilise membrane proteins in detergent based solution. PLoS One 16(7): 1–20.
- Cockcroft, S. (2021). Mammalian lipids: structure, synthesis and function. Essays Biochem. 65(5): 813–845.
- Contreras, F. X., Ernst, A. M., Wieland, F. and Brugger, B. (2011). Specificity of Intramembrane Protein-Lipid Interactions. Cold Spring Harbor Perspect. Biol. 3(6): a004705–a004705.
- Corradi, V., Sejdiu, B. I., Mesa-Galloso, H., Abdizadeh, H., Noskov, S. Y., Marrink, S. J. and Tieleman, D. P. (2019a). Emerging Diversity in Lipid–Protein Interactions. Chem. Rev. 119(9): 5775–5848.
- Corradi, V., Sejdiu, B. I., Mesa-Galloso, H., Abdizadeh, H., Noskov, S. Y., Marrink, S. J. and Tieleman, D. P. (2019b). Emerging Diversity in Lipid–Protein Interactions. Chem. Rev. 119(9): 5775–5848.
- Dörr, J. M., Scheidelaar, S., Koorengevel, M. C., Dominguez, J. J., Schäfer, M., van Walree, C. A. and Killian, J. A. (2016). The styrene–maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J. 45(1): 3–21.
- Engelman, D. M. (2005). Membranes are more mosaic than fluid. Nature 438(7068): 578–580.
- Ernst, R., Ejsing, C. S. and Antonny, B. (2016). Homeoviscous Adaptation and the Regulation of Membrane Lipids. J. Mol. Biol. 428(24): 4776–4791.
- Fahy, E., Subramaniam, S., Murphy, R. C., Nishijima, M., Raetz, C. R., Shimizu, T., Spener, F., van Meer, G., Wakelam, M. J., Dennis, E. A., et al. (2009). Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50: S9–S14.
- Horvath, S. E. and Daum, G. (2013). Lipids of mitochondria. Prog. Lipid Res. 52(4): 590-614.
- Liebisch, G., Fahy, E., Aoki, J., Dennis, E. A., Durand, T., Ejsing, C. S., Fedorova, M., Feussner, I., Griffiths, W. J., Köfeler, H., et al. (2020). Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 61(12): 1539–1555.
- Merklinger, L., Bauer, J., Pedersen, P. A., Damgaard, R. B. and Morth, J. P. (2022). Phospholipids alter activity and stability of mitochondrial membrane-bound ubiquitin ligase MARCH5. Life Sci. Alliance 5(8): 1–13.
- Pettersen, J. M., Yang, Y. and Robinson, A. S. (2023). Advances in nanodisc platforms for membrane protein purification. Trends Biotechnol. 41(8): 1041–1054.
- Pye, V. E., Rosa, A., Bertelli, C., Struwe, W. B., Maslen, S. L., Corey, R., Liko, I., Hassall, M., Mattiuzzo, G., Ballandras-Colas, A., et al. (2020). A bipartite structural organization defines the SERINC family of HIV-1 restriction factors. Nat. Struct. Mol. Biol. 27(1): 78–83.
- Stangl, M. and Schneider, D. (2015). Functional competition within a membrane: Lipid recognition vs. transmembrane helix oligomerization. Biochim. Biophys. Acta Biomembr. 1848(9): 1886–1896.
- Sych, T., Levental, K. R. and Sezgin, E. (2022). Lipid–Protein Interactions in Plasma Membrane Organization and Function. Annu. Rev. Biophys. 51(1): 135–156.
- van Meer, G., Voelker, D. R. and Feigenson, G. W. (2008). Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9(2): 112–124.
- Verchère, A., Broutin, I. and Picard, M. (2017). Reconstitution of Membrane Proteins in Liposomes. Methods Mol. Biol. 259–282.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Merklinger, L. and Morth, J. P. (2023). Phospholipid Preparations to Characterize Protein–Lipid Interactions In Vitro. Bio-protocol 13(22): e4887. DOI: 10.21769/BioProtoc.4887.
Category
Biochemistry > Lipid > Lipid-protein interaction
Molecular Biology > Protein > Stability
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link