- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Differentiation of Bone Marrow Monocytes into Alveolar Macrophages-like Cells through Co-culture with Lung Epithelial Cells and Group 2 Innate Lymphoid Cells
(*contributed equally to this work) Published: Vol 13, Iss 18, Sep 20, 2023 DOI: 10.21769/BioProtoc.4818 Views: 2471
Reviewed by: Rajesh RanjanDebashis DuttaThirupugal Govindarajan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Assessing Human Treg Suppression at Single-Cell Resolution Using Mass Cytometry
Jonas Nørskov Søndergaard [...] James B. Wing
Aug 20, 2025 2923 Views

Isolation and Co-culture of Paneth Cells and Intestinal Stem Cells
Ryosuke Isotani [...] Toshimasa Yamauchi
Sep 20, 2025 3472 Views

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2243 Views
Abstract
During life, the embryonic alveolar macrophage (AM) population undergoes successive waves of depletion and replenishment in response to infectious and inflammatory episodes. While resident AMs are traditionally described as from embryonic origin, their ontogeny following inflammation or infection is much more complex. Indeed, it appears that the contribution of monocytes (MOs) to the AM pool is variable and depends on the type of inflammation, its severity, and the signals released in the microenvironment of the pulmonary niche (peripheral imprinting) and/or in the bone marrow (central imprinting). Deciphering the cellular and molecular mechanisms regulating the differentiation of MOs into AMs remains an area of intense investigation, as this could potentially explain part of the inter-individual susceptibility to respiratory immunopathologies. Here, we detail a relevant ex vivo co-culture model to investigate how lung epithelial cells (ECs) and group 2 lung innate lymphoid cells (ILC2s) contribute to the differentiation of recruited MOs into AMs. Interestingly, the presence of lung ILC2s and ECs provides the necessary niche signals to ensure the differentiation of bone marrow MOs into AMs, thus establishing an accessible model to study the underlying mechanisms following different infection or inflammation processes.
Key features
• Ex vivo co-culture model of the alveolar niche.
• Deciphering the particular niche signals underlying the differentiation of MO into AMs and their functional polarization.
Graphical overview
This protocol described the isolation of bone marrow monocytes (MOs), lung epithelial cells (ECs), and lung group 2 lung innate lymphoid cells (ILC2s) and the ex vivo co-culture of these cells to drive the differentiation of bone marrow MOs into alveolar macrophages (AMs).
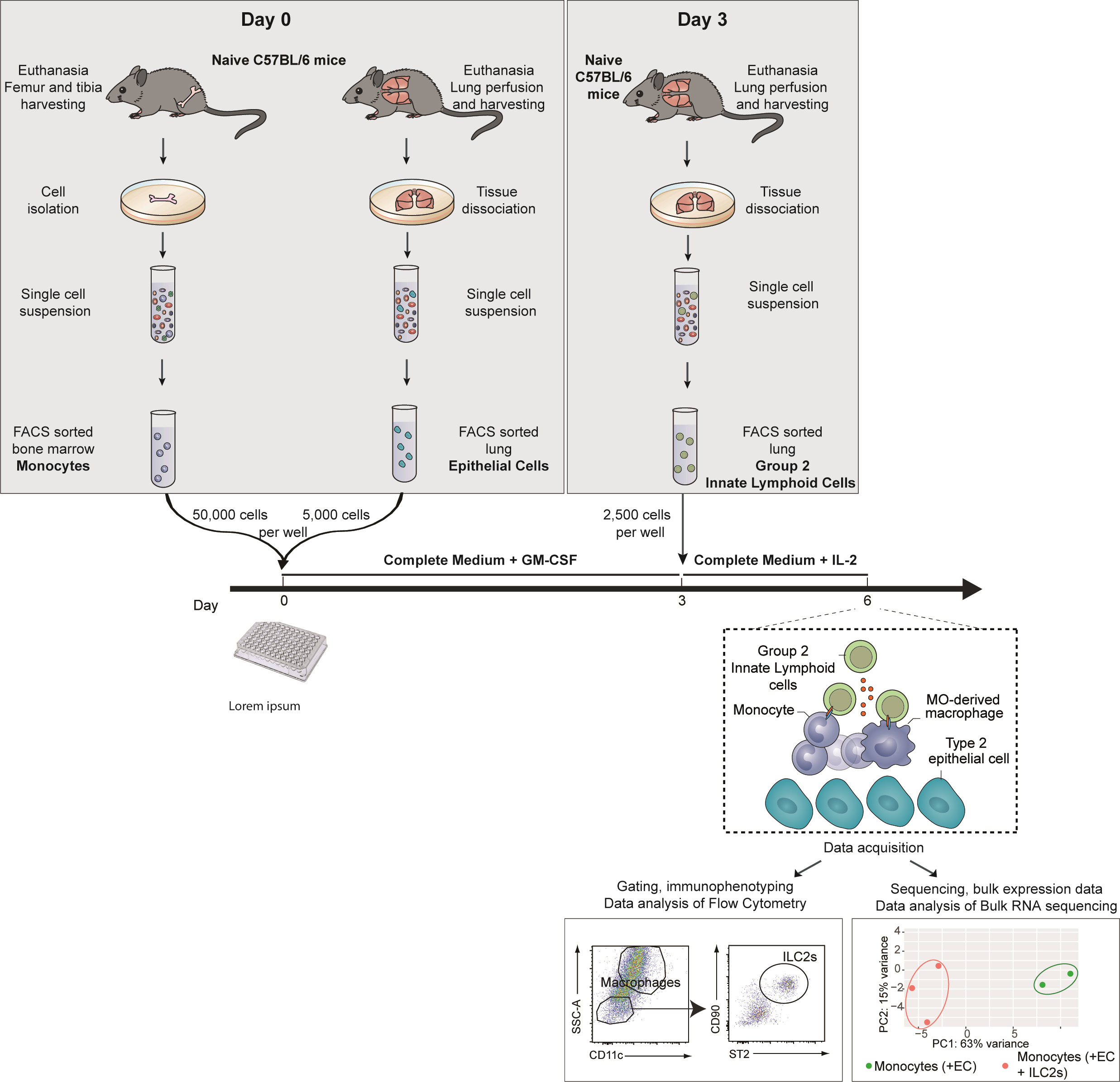
This co-culture experiment is composed of three steps (Graphical overview):
1. Identification and FACS-sorting of ECs and MOs isolated from the lung and the bone marrow of naive mice, respectively.
2. Culture of these ECs and bone marrow MOs for three days.
3. Addition of ILC2s isolated from the lung of naïve mice or mice subjected to a treatment/infection of interest.
Background
Innate lymphoid cells (ILCs) were firstly described in 2010 and have since been described as important regulators of both inflammation and homeostasis throughout the body (Moro et al., 2010; Ebbo et al., 2017). They affect the function of their neighboring cells through the cytokines they produce and also via direct cell–cell interactions (Spits and Di Santo, 2011; Oliphant et al., 2014). Their role in modulating alveolar macrophage (AMs) polarization has been described in early life through the IL-33/ILC2/IL-13 activation cascade, which confers an M2 phenotype to AMs (Saluzzo et al., 2017). In the context of a persistent viral infection, we recently highlighted the essential role of ILC2s as lung niche cells that imprint the tissue-specific identity of monocytes (MOs)-derived AMs and shape their function (Loos et al., 2023).
The protocol presented here details the optimal settings to recapitulate an alveolar niche ex vivo allowing the differentiation of MOs into AMs. In particular, this protocol describes the experimental conditions for co-culture of epithelial cells (ECs) and bone marrow MOs, with group 2 lung innate lymphoid cells (ILC2s) isolated from mouse lung. In the related publication, we showed that bone marrow MOs differentiate in vitro into AM-like cells in the presence of ILC2s and ECs, as evidenced by the expression of genes related to macrophage differentiation and activation (Csf1, PPARg, and Il4ra) (Loos et al., 2023). In addition, we also showed the long-term effect of lung gammaherpesvirus infection on ILC2s and the subsequent consequences on ILC2-mediated differentiation of MOs into AMs. In particular, we highlighted the key role of granulocyte-macrophage colony-stimulating factor (GM-CSF) produced by ILC2s in the differentiation of MO-AMs. The main advantage of this protocol is that it reduces the cell types involved in the differentiation of MO-AMs to a number of selected candidates that we can study ex vivo. This approach therefore allows to test the influence of either environmental conditions (such as viruses or pollutants) or defined mediators in a very controlled environment with every other condition being equal. Subsequently, the differentiation of MOs into AM-like cells can be analyzed classically by multiparametric flow cytometry or by transcriptomic analyses such as bulk or single-cell RNA sequencing. Besides, modifications in intracellular metabolism driving epigenetic reprogramming could be investigated via metabolic (Cedex BioAnalyzer) and epigenetic analysis (ATAC-seq, CHIP-seq). Subsequently, those differences could be confirmed using specific neutralizing antibodies or metabolic (OXPHOS inhibitor, such as oligomycin, or glycolysis manipulator, such as PKM2-pyruvate) (Surace et al., 2021) or epigenetic (BET bromodomain inhibitor IBET151) inhibitors (Kerscher et al., 2019). Altogether, this protocol presents a method to study the different aspects of the biology of MO-derived AMs under controlled and reproducible experimental conditions.
Materials and reagents
Biological materials
Female C57BL/6 mice (8 weeks old) (Charles River, C57BL/6NCrl, catalog number: 000664)
Reagents
Dulbecco’s modified Eagle medium (DMEM, high glucose, GlutaMAX Supplement, pyruvate) (Thermo Fisher Scientific, Gibco, catalog number: 31966021)
RPMI 1640 medium (Thermo Fisher, catalog number: 11875093)
Minimum essential medium (MEM) (Sigma, catalog number: M0446-500ML)
Bovine serum albumin (BSA) (Sigma, catalog number: A8412)
Fetal bovine serum (FBS) (Gibco, catalog number: 10082147)
L-Glutamine (Corning, catalog number: 25005CI)
Sodium pyruvate (Corning, catalog number: 25-000-CIR)
Penicillin-streptomycin (Gibco, catalog number: 15070063)
β-mercaptoethanol (Sigma-Aldrich, catalog number: M3148)
Phosphate-buffered saline, pH 7.4 (Gibco, catalog number: 10010-023)
EDTA (Corning, catalog number: 46-034-CI)
Dispase (Sigma-Aldrich, catalog number: 42613-33-2)
Liberase research grade (Sigma-Aldrich, catalog number: 5401119001)
DNase (Sigma-Aldrich, catalog number: 11284932001)
GentleMACS C tube (Miltenyi, catalog number: 130-093-237)
Red blood cells (RBC) lysis buffer (Thermo Fisher Scientific, catalog number: 00433357)
MojoSort mouse anti-CD45 nanobeads (BioLegend, catalog number: 480027)
MojoSort mouse anti-APC nanobeads (BioLegend, catalog number: 480072)
Recombinant murine GM-CSF (Peprotech, catalog number: 315-03)
Recombinant murine IL-2 (Peprotech, catalog number: 212-12)
Zombie Aqua Fixable Viability kit (BioLegend, catalog number: 423101)
Sytox Blue Dead Cell stain (Thermo Fisher, catalog number: s34857)
Anti-mouse CD16/32 Fc block clone 93 (BioLegend, catalog number: 101301)
Paraformaldehyde (PFA) (Santa Cruz Biotechnology, catalog number: sc-281692)
Saponin (Sigma-Aldrich, catalog number: 47036)
Sodium azide (NaN3) (Sigma-Aldrich, catalog number: S2002)
TRIzol reagent (Thermo Fisher, catalog number: 15596026)
Flow cytometry antibodies
Antibodies for bone marrow MO enrichment (bead-based negative selection) and sorting:
Negative selection
i. Anti-mouse B220/CD45R APC (clone RA3-6B2) (BioLegend, catalog number: 103212)
ii. Anti-mouse Ly6G APC (clone 1A8) (BioLegend, catalog number: 127613)
Flow cytometry mastermix staining
i. Anti-mouse CD11b BV711 (clone M1/70) (BD Biosciences, catalog number: 563168)
ii. Anti-mouse Ly6G APC-Cy7 (clone 1A8) (BD Biosciences catalog number: 560600)
iii. Anti-mouse Ly6C BV785 (clone HK1.4) (BioLegend, catalog number: 128041)
iv. Anti-mouse I-A/I-E PE/Cyanine7 (clone M5/114.15.2) (BioLegend, catalog number: 107630)
v. Anti-mouse CD3e FITC (clone 145-2C11) (BioLegend, catalog number: 100306)
Antibodies for lung EC enrichment (CD45 bead-based negative selection) and sorting:
Flow cytometry mastermix staining
i. Anti-mouse CD45 PE/Cyanine7 (clone 30-F11) (BioLegend, catalog number: 103114)
ii. Anti-mouse EpCAM FITC (clone G8.8) (BioLegend, catalog number: 118207)
iii. Anti-mouse CD31 PE (clone 390) (BioLegend, catalog number: 102407)
Antibodies for lung ILC2 enrichment (bead-based negative selection) and sorting:
Negative selection
i. Anti-mouse B220/CD45R APC (clone RA3-6B2) (BioLegend, catalog number: 103212)
ii. Anti-mouse CD11c APC (clone N418) (BioLegend, catalog number: 117310)
iii. Anti-mouse CD3e APC (clone 145-2C11) (BioLegend, catalog number: 100312)
iv. Anti-mouse CD4 APC (clone RM 4-5) (BioLegend, catalog number: 100515)
v. Anti-mouse CD49b APC (clone DX5) (BioLegend, catalog number: 108910)
vi. Anti-mouse CD5 APC (clone 53-7.3) (BioLegend, catalog number: 100626)
vii. Anti-mouse CD8a APC (clone 53-6.7) (BioLegend, catalog number: 100711)
viii. Anti-mouse F4/80 APC (clone BM8) (BioLegend, catalog number: 123116)
ix. Anti-mouse FcϵRIα APC (clone MAR-1) (BioLegend, catalog number: 134316)
x. Anti-mouse Gr-1 APC (clone RB6-8C5) (BioLegend, catalog number: 108412)
xi. Anti-mouse Siglec-F APC (clone E50-2440) (BioLegend, catalog number: 155507)
Flow cytometry mastermix staining
i. Anti-mouse CD45 PE/Cyanine7 (clone 30-F11) (BioLegend, catalog number: 103114)
ii. Anti-mouse B220/CD45R APC (clone RA3-6B2) (BioLegend, catalog number: 103212)
iii. Anti-mouse CD11c APC (clone N418) (BioLegend, catalog number: 117310)
iv. Anti-mouse CD3e APC (clone 145-2C11) (BioLegend, catalog number: 100312)
v. Anti-mouse CD4 APC (clone RM 4-5) (BioLegend, catalog number: 100515
vi. Anti-mouse CD49b APC (clone DX5) (BioLegend, catalog number: 108910
vii. Anti-mouse CD5 APC (clone 53-7.3) (BioLegend, catalog number: 100626
viii. Anti-mouse CD8a APC (clone 53-6.7) (BioLegend, catalog number: 100711
ix. Anti-mouse F4/80 APC (clone BM8) (BioLegend, catalog number: 123116)
x. Anti-mouse FcϵRIα APC (clone MAR-1) (BioLegend, catalog number: 134316)
xi. Anti-mouse Gr-1 APC (clone RB6-8C5) (BioLegend, catalog number: 108412)
xii. Anti-mouse Siglec-F APC (clone E50-2440) (BioLegend, catalog number: 155507)
xiii. Anti-mouse CD25 Alexa Fluor 700 (clone PC61) (BioLegend, catalog number: 102024)
xiv. Anti-mouse ST2 PE (clone DIH9) (BioLegend, catalog number: 145304)
xv. Anti-mouse CD90.2 BV711 (clone 53-2.1) (BD Biosciences, catalog number: 740647
Solutions
Wash buffer (see Recipes)
MACS buffer (see Recipes)
FACS buffer (see Recipes)
Complete culture medium (see Recipes)
Lung digestion solution (see Recipes)
MO and EC culture medium (see Recipes)
ILC2 culture medium (see Recipes)
Permeabilization buffer (see Recipes)
Intracellular staining buffer (see Recipes)
Recipes
Wash buffer
PBS-EDTA 2 mM with 10% FBS
MACS buffer
PBS containing 0.5% BSA and 2 mM EDTA
Keep buffer cold (2–8 °C)
FACS buffer
PBS containing 0.5% BSA, 2 mM EDTA, 2 mM sodium azide (NaN3)
Complete culture medium
RPMI medium supplemented with 10% FBS, 1% MEM, 100 U/mL penicillin, 100 U/mL streptomycin, 2 mM L-glutamine, 50 μM of β-mercaptoethanol
Lung digestion solution
Freshly prepare 1.5 mL/sample of RPMI containing 50 μg/mL Liberase and 100 μg/mL DNase I
MO and EC culture medium
RPMI medium supplemented with 10% FBS, 1% MEM, 100 U/mL penicillin, 100 U/mL streptomycin, 2 mM L-glutamine, 50 μM of β-mercaptoethanol, and 10 ng/mL recombinant murine GM-CSF
ILC2 culture medium
RPMI-1640 medium supplemented with 10% FBS, 1% MEM, 100 U/mL penicillin, 100 U/mL streptomycin, 2 mM L-glutamine, 50 μM of β-mercaptoethanol, and 10 ng/mL recombinant mouse IL-2
Permeabilization buffer
PBS containing 0.1% of saponin
Keep buffer cold and sterile after filtration
Intracellular staining buffer
FACS buffer (see Recipe 3) containing 0.1% of saponin
Laboratory supplies
15 and 50 mL conical centrifuge tubes (Corning, catalog numbers: 352096 and 352070)
Cell culture 60 × 15 mm Petri dishes (Thermo Fisher Scientific, catalog number: 150288)
24-well cell culture plates (Falcon, catalog number: 353047)
96-well round (U) bottom plate (Thermo Scientific, catalog number: 163320)
10 mL syringes (Terumo, catalog number: SS+10ES1)
25 G needles (Terumo, catalog number: AN*2516R1)
Falcon® 70 μm cell strainer (Corning, catalog number: 352350)
LD columns (Miltenyi, catalog number: 130-042-901)
Equipment
Swing-bucket centrifuge (Eppendorf, model: 5810R)
Tabletop centrifuge (Eppendorf, model: 5427R)
Inverted light microscope (NIKON, Eclipse TS100)
Fluorescence-activated cell sorter (BD Biosciences, model: Aria IIIu)
Flow cytometer (BD LSR Fortessa X-20)
Laminar flow hood (FASTER SafeFast Classic)
Scalpel (Swann-Morton 210 mm No.3 ref 0933)
Forceps (Bochem Stainless steel 18/10 ref 1141)
GentleMACS dissociator (Miltenyi)
QuadroMACS Separators (Miltenyi)
Hemocytometer (Marienfeld, 0640710)
Software and datasets
FlowJo v10 (FlowJo LLC)
FACSDiVa software
Procedure
In order to get enough lung ECs and ILC2s to perform the co-culture settings, purify lung ECs and ILC2s from five mice. This will allow you to obtain, after each enrichment, 12–15 wells of 5 × 103 lung ECs and 2.5 × 103 ILC2s.
Caution: The entire procedure must be performed under a laminar flow hood to maintain the culture sterile.
Bone marrow MOs isolation
Preparing bone marrow cell suspension
Euthanize the mice and spray the skin with 75% ethanol.
Remove the skin from the lower part of the body.
Remove tissues from legs with scissors.
Separate the femur from the femoral capsule. Be careful not to break the bones; the best is to dislocate the bones rather than cutting them. Place femur and tibia of one leg into a 50 mL tube filled with 20 mL of DMEM on ice.
Take the bones and cut off both ends with a scalpel in a Petri dish.
Using a 10 mL syringe with a 25 G needle, flush the bone marrow cells into a 50 mL tube with wash buffer (see Recipes).
Pipette to obtain a homogeneous cell suspension and filter it through a 70 μm cell strainer into a new 50 mL tube.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant.
Resuspend in 1 mL of RBC lysis buffer. Incubate for 5 min at room temperature (RT) and add 9 mL of wash buffer. Count cells using the hemocytometer.
Note: You should expect approximately 40 × 106–60 × 106 cells with ~94%–98% viability for bone marrow cell collection from both femurs and tibias of an uninfected 8-week-old female C57BL/6 mouse.
Negative selection
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant.
Resuspend in MACS buffer (108 cells/mL) (see Recipes) containing anti-Ly6G and B220 antibodies coupled with APC fluorochrome (see Flow cytometry antibodies 1.a.i) and incubate for 20 min at 4 °C in the dark.
Note: The B220 and Ly6G antibodies target B cells and neutrophils, respectively, that will be further depleted, thereby facilitating FACS sorting.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend in 1 mL of ice-cold MACS buffer containing purified anti-CD16/32 antibody (diluted 1:500) to block Fc receptors.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant.
Resuspend the cells in MACS buffer (107 cells/100 μL) with the adapted volume of MojoSort APC Nanobeads (10 μL/107 cells) (according to MojoSort Mouse anti-APC Nanobeads protocol). Incubate on ice for 15 min.
Wash the cells by adding MACS buffer up to 4 mL. Centrifuge the cells at 250× g for 5 min at 4 °C. Discard the supernatant.
Resuspend in MACS buffer (5 × 107 cells/500 μL).
Filter cells through 70 μm cell strainer.
Proceed with magnetic negative selection: apply cell suspension onto the prepared LD column (according to Miltenyi LD column protocol). Collect flowthrough containing unlabeled cells after washes with 2 × 1 mL of MACS buffer. The unlabeled cells contain MOs.
MO staining for FACS sorting
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend the pellet in FACS buffer (107 cells/mL) (see Recipes) containing antibodies (anti-CD19, CD3, Ly6G, B220, CD11b, Ly6C) for 20 min at 4 °C in the dark. Centrifuge the cells at 250× g for 5 min at 4 °C. Discard the supernatant. Resuspend the pellet in 5 mL of PBS and repeat centrifugation.
Discard supernatant and perform viability staining by adding SYTOX Blue to FACS buffer (1 μL/10 mL/5 × 107 cells). Incubate for 5 min at RT in the dark.
Perform FACS sorting using FACS Aria IIIu. Bone marrow MOs are gated as Ly6G-MHCII-CD3-CD11b+Ly6C+ living cells (Figure 1).
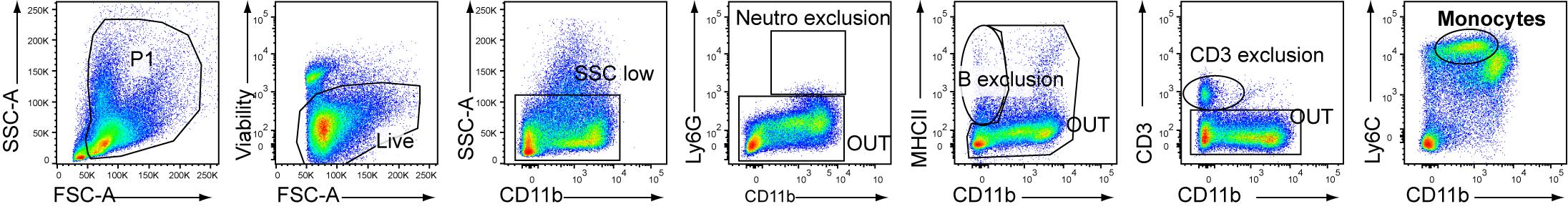
Figure 1. Gating strategy for FACS sorting of bone marrow monocytes (MOs), described as SSC-AlowLy6C+CD11b+ living cells after exclusion of neutrophils (Ly6G+CD11b+), B cells (MHCII+CD11b-), and T cells (CD3+CD11b-) after cell enrichment through MACS-negative selectionCount the cells using the hemocytometer.
Centrifuge at 250× g for 5 min at 4 °C. Discard supernatant and resuspend the cells at 105 cells/100 μL of complete culture medium (see Recipes).
Lung ECs isolation
Preparing lung cell suspension
Euthanize the mice and spray the skin with 75% ethanol.
Catheterize the trachea and add 1 mL of PBS with 10 U/mL of Dispase.
Perfuse through the right ventricle with 5 mL of ice-cold PBS. Dissociate lobes of lung and place them into a 24-well plate with 2 mL of DMEM medium with 10 U/mL of Dispase for 10 min at RT.
Transfer the lung lobes from one mouse into an individual GentleMACS tube containing the lung digestion solution (see Recipes) and process with a gentleMACS dissociator (lung program 01); finally, incubate for 30 min at 37 °C. After digestion, run the lung program 02 with the gentleMACS dissociator.
Filter the cell suspension through a 70 μm cell strainer into a 50 mL tube and top up to 25 mL with wash buffer.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend the pellet in 1 mL of RBC lysis buffer. Incubate for 5 min at RT, add 9 mL of wash buffer, and pool all the samples into one 50 mL tube. Count cells using the hemocytometer.
Note: You should expect approximately 6 × 106–12 × 106 cells per lung of a naïve 8-week-old female C57BL/6 mouse.
Negative selection
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend in MACS buffer (107 cells/100 μL) with the adapted volume of MojoSort mouse CD45 nanobeads (10 μL/107 cells) according to the manufacturer protocol. Incubate for 15 min at 4 °C.
Wash the cells by adding MACS buffer up to 4 mL. Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant.
Resuspend in MACS buffer (5 × 107 cells/500 μL).
Filter cells through 70 μm cell strainer.
Proceed with magnetic negative selection: apply the cell suspension onto the prepared LD column (follow the Miltenyi LD column protocol). Collect flowthrough containing unlabeled cells after washes with 2 × 1 mL of MACS buffer.
Note: The unlabeled cells contain the non-immune cells.
ECs staining for FACS sorting
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend in FACS buffer (107 cells/mL) with antibodies (anti-CD45, CD31, EpCAM) for 20 min at 4 °C in the dark.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend the pellet in 5 mL of PBS and repeat centrifugation.
Discard supernatant and perform viability staining by adding SYTOX Blue in FACS buffer (1 μL/1 mL/5 × 107 cells). Incubate for 5 min at RT in the dark.
Proceed to FACS sorting. Lung ECs are gated as CD45-CD31-Epcam+ living cells (Figure 2). Count the cells using the hemocytometer.
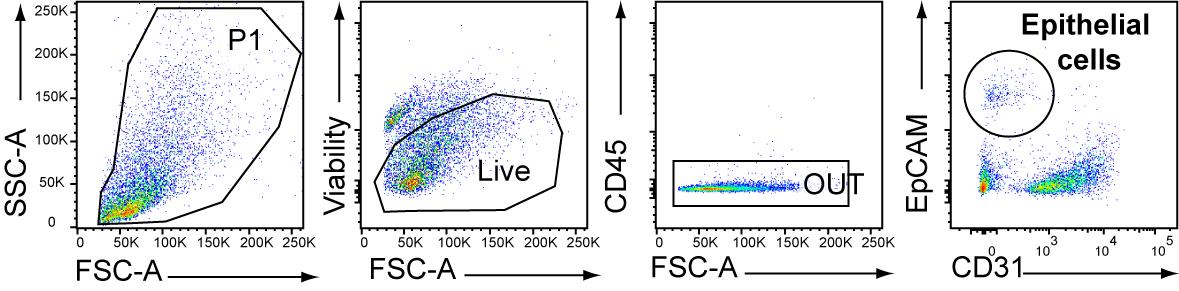
Figure 2. Gating strategy for FACS sorting of lung epithelial cells (ECs), described as CD45-CD31-EpCAM+ living cells after cell enrichment though MACS-negative selection (CD45).Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend the cells at 5 × 103 cells/50 μL in complete culture medium (see Recipes).
Culture of MOs and lung ECs
Note: The presence of lung ECs with bone marrow MOs will help to obtain MOs in the process of differentiation into AMs.
Add 50 μL (5 × 104 cells) of bone marrow MOs and 50 μL (5 × 103 cells) of lung ECs into a well of a 96-well round-bottom plate.
Add 100 μL of complete culture medium containing 20 ng/mL recombinant murine GM-CSF (for a final concentration of 10 ng/mL) into each well and gently resuspend. Caution: Provide control wells without GM-CSF as well as conditions without ECs.
Incubate at 37 °C with 5% CO2 for three days.
Lung ILC2s isolation
Preparing lung cell suspension
Euthanize the mice and spray the skin with 75% ethanol.
Perfuse through the right ventricle with 5 mL of ice-cold PBS. Transfer the lung lobes into a GentleMACS tube containing lung digestion solution (see Recipes) and process with a gentleMACS dissociator (lung program 01); finally, incubate for 30 min at 37 °C. After digestion, run the lung program 02 with the gentleMACS dissociator. Filter the cell suspension through a 70 μm cell strainer into a 50 mL Falcon tube and top up to 25 mL with wash buffer.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend the pellet in 1 mL of RBC lysis buffer. Incubate for 5 min at RT and add 9 mL of wash buffer and pool five samples at a time into one 50 mL Falcon tube. Count cells using the hemocytometer.
Note: You should expect approximately 6 × 106–10 × 106 total cells per lung of a naïve 8-week-old female C57BL/6 mouse.
Negative selection
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend the pellet at 108 cells/mL of MACS buffer with lineage marker antibodies (anti-B220, CD11c, CD3, CD4, CD5, CD8α, F4/80, FcϵR1, CD49b, and Siglec-F) conjugated to APC fluorochrome, for 20 min at 4 °C in the dark.
Note: ILC2s represent approximately 1% of the lung leukocytes population; the lineage will target other immune cells such as T cells, B cells, NK cells, and macrophages, and then will improve the FACS sorting.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend the pellet in 5 mL of MACS buffer and repeat centrifugation.
Discard the supernatant and resuspend in MACS buffer (107 cells/100 μL) with the adapted volume of MojoSort APC nanobeads (10 μL/107 cells) (follow manufacturer’s protocol). Incubate on ice for 15 min.
Wash the cells by adding MACS buffer up to 4 mL. Centrifuge the cells at 250× g for 5 min at 4 °C. Discard the supernatant.
Resuspend in MACS buffer (5 × 107 cells/500 μL).
Filter cells through a 70 μm cell strainer.
Proceed with magnetic negative selection: apply cell suspension onto the prepared LD column (follow the Miltenyi LD columns protocol). Collect flowthrough containing unlabeled cells after washes with 2 × 1 mL of MACS buffer.
Caution: The unlabeled cells contain lung ILCs and non-immune cells.
ILC2s staining for FACS sorting
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend in FACS buffer (107 cells/mL) with antibodies (anti-B220, CD11c, CD3, CD4, CD5, CD8α, F4/80, FcϵR1, Ly6G, Siglec-F, CD45, CD90.2, ST2, and CD25) for 20 min at 4 °C in the dark. Centrifuge the cells at 250× g for 5 min at 4 °C. Discard the supernatant. Resuspend the pellet in 5 mL of PBS and repeat centrifugation.
Discard supernatant and perform viability staining by adding SYTOX Blue to FACS buffer (1 μL/1 mL/5 × 107 cells). Incubate for 5 min at RT in the dark.
Proceed to FACS sorting using FACS Aria IIIu following manufacturer’s protocol. Lung ILC2s are gated as CD45+Lin-CD90.2+ST2+CD25+ living cells (Figure 3). Count cells using the hemocytometer.
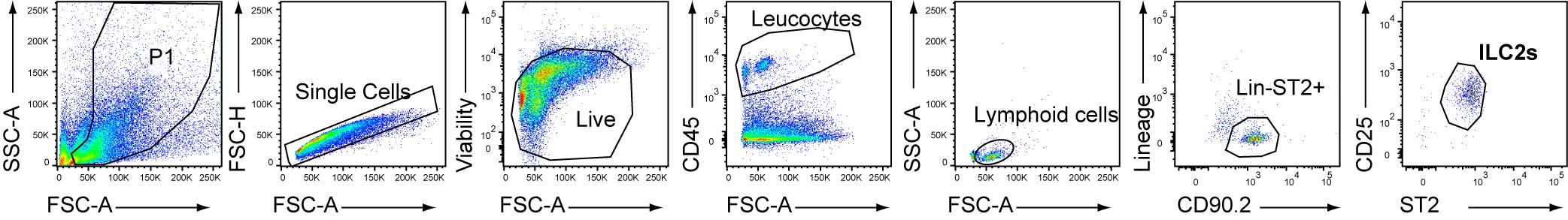
Figure 3. Gating strategy for FACS sorting of lung group 2 lung innate lymphoid cells (ILC2s) after cell enrichment through MACS-negative selection against lineage markers (B220, CD11c, CD3, CD4, CD49b, CD5, CD8α, F4/80, FcϵR1, Gr1 and Siglec-F). ILC2s were identified as Lin-CD45+ST2+CD90.2+CD25+.Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend cells at 2.5 × 103 cells/200 μL of ILC2 culture medium (see Recipes). Critical: IL-2 is mandatory to maintain ILC2s in ex vivo culture.
Co-culture of ILC2s with MOs and ECs
This step allows to investigate the role of interactions between ILC2s and MOs differentiating into AMs and to study the potential importance of a targeted ligand–receptor interaction.
Centrifuge the 96-well plate with the bone marrow MOs and lung ECs co-culture at 250× g for 5 min at 4 °C. Gently aspirate the supernatant without touching the pellet.
Add 200 μL of ILC2s (2.5 × 103 cells) in the ILC2 culture medium. Gently mix the cells together. Incubate at 37 °C in 5% CO2 environment for three days.
Caution: Provide control conditions without ILC2s.
Harvest and staining for FACS analysis
Note: A substantial fraction of bone marrow MOs differentiate into AM-like cells, as observed by the expression of CD11c. MO-derived AMs acquire the M2 marker Arg1 when co-cultured with ILC2s from a naïve mouse.
After three days, centrifuge the plate at 250× g for 5 min at 4 °C.
Perform viability staining by resuspending cell pellets in 100 μL/well in PBS with 1:1,500 Zombie AquaTM for 15 min at RT in the dark. Critical: Use fixable viability kit if the assessment requires intracytoplasmic staining such as Arg1.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant.
Resuspend cell pellets in 50 μL/well in FACS buffer with the following antibodies (anti-CD45, CD11c, Siglec-F, MHC-II, CD90, ST2) or any antibodies of targets of your interest, for 20 min at 4 °C in the dark.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend in 100 μL/well of PFA 2% for at least 15 min at 4 °C in the dark (it may be extended overnight).
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend in 100 μL/well of permeabilization buffer (see Recipes) and incubate at 37 °C for 15 min.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend in 100 μL/well of intracellular staining buffer (see Recipes) with antibody against intracellular antigens. Incubate for 30 min at RT in the dark.
Centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend in 200 μL/well of FACS buffer and repeat the centrifugation.
Discard the supernatant, resuspend in 200 μL/well of FACS buffer, and proceed to FACS analysis using a BD LSR Fortessa X-20 (Figure 4).
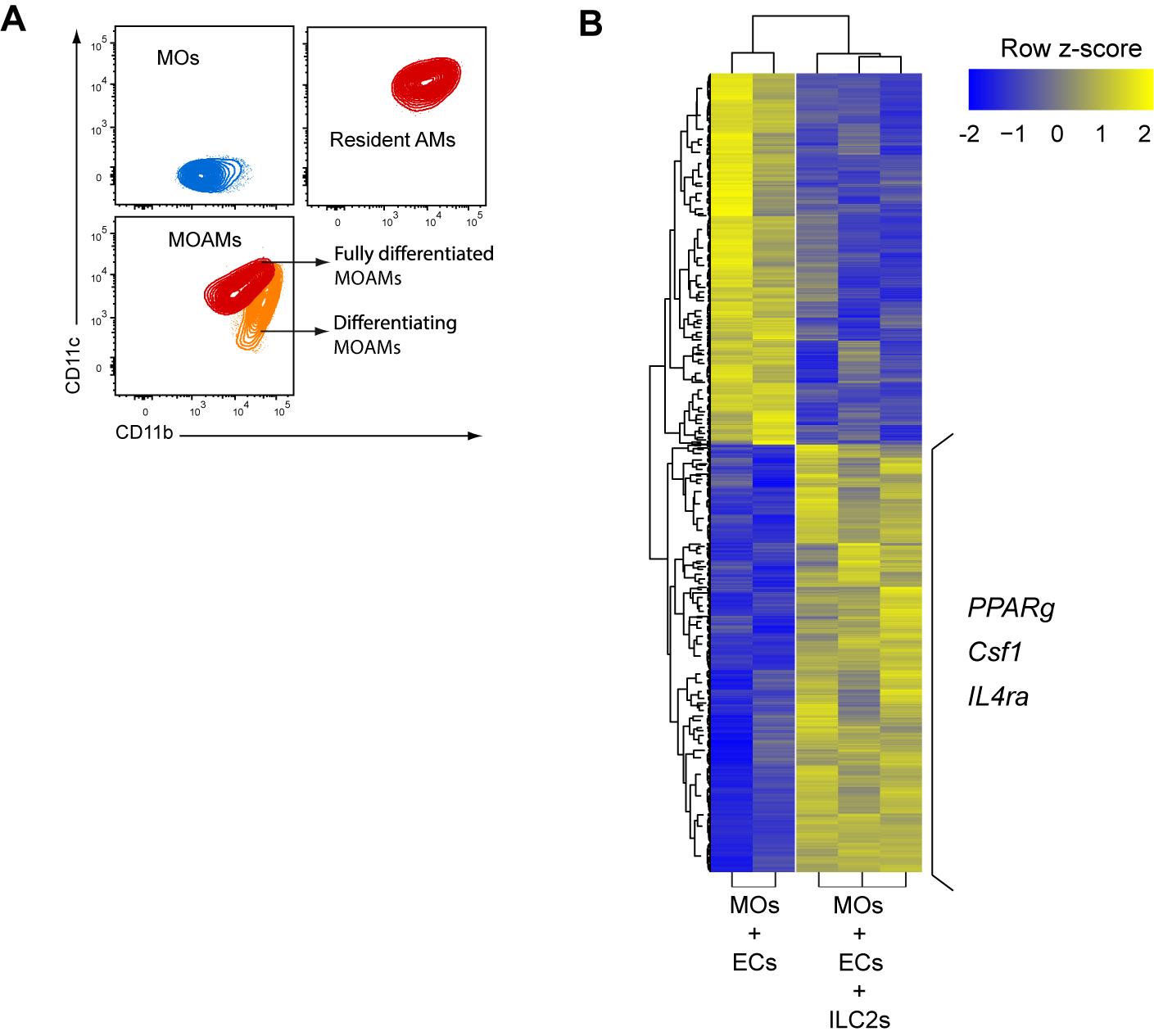
Figure 4. Lung ILC2s regulate differentiation and prone a M2 phenotype of MOAMs in vitro. (A) Representative flow cytometry plots of bone marrow monocytes (MOs), bronchoalveolar lavage alveolar macrophage (AMs) and bone marrow MOs differentiated to MOAMs from co-culture with lung epithelial cells (ECs) and lung group 2 lung innate lymphoid cells (ILC2s). (B) Heatmap of differentially expressed genes (False Discovery Rate, FRD) < 0.05; change in expression of over two-fold) in MO-derived macrophages isolated from co-cultures of MOs and ECs with or without ILC2s. Three genes with higher expression in the condition with ILC2s are highlighted.Harvest for RNA sequencing
Note: For a deeper analysis, transcriptomic analysis can be performed on the ex vivo differentiated MO-derived AMs.
After step E1, centrifuge at 250× g for 5 min at 4 °C and discard the supernatant. Resuspend in 100 μL of FACS buffer with antibodies (anti-Ly6G, Ly6C, CD11c, CD45, CD90.2, ST2, and CD25) for 20 min at 4 °C in the dark. Centrifuge the cells at 250× g for 5 min at 4 °C. Discard the supernatant. Resuspend the pellet in 5 mL of PBS and repeat centrifugation.
Discard supernatant and perform viability staining by adding SYTOX Blue in FACS buffer (1 μL/1 mL/5 × 107 cells/). Incubate for 5 min at RT in the dark.
Proceed to FACS sorting using FACS Aria IIIu by following manufacturer’s protocol. MO-derived AMs are defined as Ly6G-, Ly6C-, autofluorescent, and CD11c+ living cells, and directly sorted in 1 mL of TRIzol before being snap frozen until future analysis (Figure 4).
Data analysis
Samples were sorted on a BD FACS Aria IIIu using FACSDiVa software. Flow cytometry data were analyzed using FlowJo to gate, quantify, and analyze the different populations.
Cells were sorted based on the expression of specific markers:
• Bone marrow MOs were gated as Ly6G-MHCII-CD3-CD11b+Ly6C+ living cells (Figure 1).
• Lung ECs were gated as CD45-CD31-Epcam+ living cells (Figure 2)
• Lung ILC2s were gated as CD45+Lin-CD90.2+ST2+CD25+ living cells (Figure 3).
For the readout, MO-derived AMs were defined as Ly6G-, Ly6C-, autofluorescent, and CD11c+ living cells, and ILC2s as SSC-AlowCD11c-CD90.2+ST2+ living cells.
Detailed information about data analyses appears in the original research article (Loos et al., 2023 in Figure 6 and Figure S4).
Validation of protocol
Loos et al. (2023). Dampening type 2 properties of group 2 innate lymphoid cells by a gammaherpesvirus infection reprograms alveolar macrophages. Science Immunology (Figure 6 and Figure S4).
General notes and troubleshooting
The entire procedure must be performed under a laminar flow hood to maintain sterile conditions and avoid any contamination.
ILC2s and epithelial cells are a minority population in the lung, so a minimum of five individuals per condition should be pooled to obtain enough cells of interest in order to generate triplicates.
Bead enrichment will facilitate FACS sorting, and negative enrichment will avoid early cell activation.
As ILC2 and epithelial cells phenotypes are directly impacted by environmental conditions, the outcome of the experiment largely depends on the environmental conditions to which mice have been subjected, as seen for example with MuHV-4 infection (Loos et al., 2023).
Acknowledgments
The protocol was adapted from the previously published paper: Loos et al. (2023).
This work was supported, in part, by University of Liège (VIRIMPRINT ARC to L.G.), by Fondation Léon Fredericq (grant to P.L.), by the Fonds de la Recherche Scientifique—Fonds National Belge de la Recherche Scientifique (F.R.S./FNRS, “credit de recherche” J007515F to L.G.; “projet de recherche” T.0195.16 to L.G.; research associate support for B.M.; and research fellow for P.L. and C.M.), by Institut MERIEUX starting grant (to L.G.), by EOS joint programme of F.R.S./FNRS Fonds wetenschapellijk onderzoek–Vlaanderen-FWO (EOS ID:30981113 and 40007527) (to L.G.); and by ERC Starting Grant (ERC-StG-2020 VIROME, ID:853608) (to B.M.).
Competing interests
The authors declare no competing interests.
Ethical considerations
All work in the development of this protocol was approved by the Committee on the Ethics of Animal Experiments of ULiège (permit numbers 1845).
References
Ebbo, M., Crinier, A., Vély, F. and Vivier, E. (2017). Innate lymphoid cells: major players in inflammatory diseases. Nat. Rev. Immunol. 17(11): 665–678.
- Kerscher, B., Barlow, J. L., Rana, B. M., Jolin, H. E., Gogoi, M., Bartholomew, M. A., Jhamb, D., Pandey, A., Tough, D. F., van Oosterhout, A. J. M., et al. (2019). BET Bromodomain Inhibitor iBET151 Impedes Human ILC2 Activation and Prevents Experimental Allergic Lung Inflammation. Front. Immunol. 10: e00678.
- Loos, P., Baiwir, J., Maquet, C., Javaux, J., Sandor, R., Lallemand, F., Marichal, T., Machiels, B. and Gillet, L. (2023). Dampening type 2 properties of group 2 innate lymphoid cells by a gammaherpesvirus infection reprograms alveolar macrophages. Sci. Immunol. 8(80): eabl9041.
- Moro, K., Yamada, T., Tanabe, M., Takeuchi, T., Ikawa, T., Kawamoto, H., Furusawa, J., Ohtani, M., Fujii, H., Koyasu, S., et al. (2010). Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463(7280): 540–544.
- Oliphant, C. J., Hwang, Y. Y., Walker, J. A., Salimi, M., Wong, S. H., Brewer, J. M., Englezakis, A., Barlow, J. L., Hams, E., Scanlon, S. T., et al. (2014). MHCII-Mediated Dialog between Group 2 Innate Lymphoid Cells and CD4+ T Cells Potentiates Type 2 Immunity and Promotes Parasitic Helminth Expulsion. Immunity 41(2): 283–295.
- Saluzzo, S., Gorki, A. D., Rana, B. M., Martins, R., Scanlon, S., Starkl, P., Lakovits, K., Hladik, A., Korosec, A., Sharif, O., et al. (2017). First-Breath-Induced Type 2 Pathways Shape the Lung Immune Environment. Cell Rep. 18(8): 1893–1905.
- Surace, L., Doisne, J. M., Croft, C. A., Thaller, A., Escoll, P., Marie, S., Petrosemoli, N., Guillemot, V., Dardalhon, V., Topazio, D., et al. (2021). Dichotomous metabolic networks govern human ILC2 proliferation and function. Nat. Immunol. 22(11): 1367–1374.
- Spits, H. and Di Santo, J. P. (2011). The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12(1): 21–27.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Loos, P., Marichal, T., Machiels, B. and Gillet, L. (2023). Differentiation of Bone Marrow Monocytes into Alveolar Macrophages-like Cells through Co-culture with Lung Epithelial Cells and Group 2 Innate Lymphoid Cells. Bio-protocol 13(18): e4818. DOI: 10.21769/BioProtoc.4818.
- Loos, P., Baiwir, J., Maquet, C., Javaux, J., Sandor, R., Lallemand, F., Marichal, T., Machiels, B. and Gillet, L. (2023). Dampening type 2 properties of group 2 innate lymphoid cells by a gammaherpesvirus infection reprograms alveolar macrophages. Sci. Immunol. 8(80): eabl9041.
Category
Immunology > Immune cell isolation > Macrophage
Cell Biology > Cell isolation and culture > Co-culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









