- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
13CO2-labelling and Sampling in Algae for Flux Analysis of Photosynthetic and Central Carbon Metabolism
(*contributed equally to this work) Published: Vol 13, Iss 17, Sep 5, 2023 DOI: 10.21769/BioProtoc.4808 Views: 1919
Reviewed by: John P PhelanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
![Assessing Long-distance Transport from Photosynthetic Source Leaves to Heterotrophic Sink Organs with [<sup>14</sup>C]CO<sub>2</sub>](https://en-cdn.bio-protocol.org/imageup/arcimg/20171211011113752.jpg?t=1768618355)
Assessing Long-distance Transport from Photosynthetic Source Leaves to Heterotrophic Sink Organs with [14C]CO2
Umesh P Yadav [...] Brian G Ayre
Dec 20, 2017 6568 Views
![Quantifying the Capacity of Phloem Loading in Leaf Disks with [<sup>14</sup>C]Sucrose](https://en-cdn.bio-protocol.org/imageup/arcimg/20171211011350163.jpg?t=1768618355)
Quantifying the Capacity of Phloem Loading in Leaf Disks with [14C]Sucrose
Umesh P Yadav [...] Brian G Ayre
Dec 20, 2017 8012 Views

Quantification of Starch in Guard Cells of Arabidopsis thaliana
Sabrina Flütsch [...] Diana Santelia
Jul 5, 2018 11003 Views
Abstract
The flux in photosynthesis can be studied by performing 13CO2 pulse labelling and analysing the temporal labelling kinetics of metabolic intermediates using gas or liquid chromatography linked to mass spectrometry. Metabolic flux analysis (MFA) is the primary approach for analysing metabolic network function and quantifying intracellular metabolic fluxes. Different MFA approaches differ based on the metabolic state (steady vs. non-steady state) and the use of stable isotope tracers. The main methodology used to investigate metabolic systems is metabolite steady state associated with stable isotope labelling experiments. Specifically, in biological systems like photoautotrophic organisms, isotopic non-stationary 13C metabolic flux analysis at metabolic steady state with transient isotopic labelling (13C-INST-MFA) is required. The common requirement for metabolic steady state, alongside its very short half-timed reactions, complicates robust MFA of photosynthetic metabolism. While custom gas chambers design has addressed these challenges in various model plants, no similar tools were developed for liquid photosynthetic cultures (e.g., algae, cyanobacteria), where diffusion and equilibration of inorganic carbon species in the medium entails a new dimension of complexity. Recently, a novel tailor-made microfluidics labelling system has been introduced, supplying short 13CO2 pulses at steady state, and resolving fluxes across most photosynthetic metabolic pathways in algae. The system involves injecting algal cultures and medium containing pre-equilibrated inorganic 13C into a microfluidic mixer, followed by rapid metabolic quenching, enabling precise seconds-level label pulses. This was complemented by a 13CO2-bubbling-based open labelling system (photobioreactor), allowing long pulses (minutes–hours) required for investigating fluxes into central C metabolism and major products. This combined labelling procedure provides a comprehensive fluxome cover for most algal photosynthetic and central C metabolism pathways, thus allowing comparative flux analyses across algae and plants.
Keywords: PhotosynthesisBackground
Over the last two decades, numerous analytical and computational approaches have emerged for deciphering metabolic networks. Understanding metabolic pathways and their regulation is crucial for metabolic engineering, biotechnology, or pharmacology, which can be explored through metabolic flux analysis (MFA) studies. Metabolite levels do not provide reliable information about pathway flux, due to its complex relationship with protein activity and substrate level (Blank and Sauer, 2004). Major progress has been made through the use of isotopic labelling, allowing a better and more accurate assessment of the concentration of metabolites by NMR, gas chromatography (GC), or liquid chromatography (LC) mass spectrometry (MS) and combinations thereof (Niedenführ et al., 2015). Various tools have been developed alongside computational and mathematical modelling methods (e.g., INCA, METRAN, SUMOFLUX), aiming to fit a theoretical model to estimate metabolic fluxes (Heise et al., 2015).
Isotopic non-stationary 13C metabolic flux analysis (13C-INST-MFA) offers a robust framework for flux estimation in single carbon substrate–based systems (e.g., autotrophic, methanotrophic) (Jazmin and Young, 2013). Aiming to resolve differential balance equations for the time-dependent labelling of intermediate metabolites, 13C-INST-MFA is ideally suited to slowly labelled systems (e.g., due to large intermediate pools or pathway bottlenecks). Thus, as the entry point for the single-carbon substrate system of most photoautotrophs, photosynthetic metabolism represents a unique challenge for 13C-INST-MFA, due to its relatively small intermediate pool size and, consequently, very rapid turnover rates (Szecowka et al., 2013; Heise et al., 2014; Abernathy et al., 2017; Allen and Young, 2020). Despite this challenge, this approach has been widely used in the past decade to estimate fluxes in photoautotrophic systems, ranging from cyanobacteria to land plants (Ma et al., 2014; Adebiyi et al., 2015; Cheah and Young, 2018; Wieloch, 2021; Xu et al., 2022).
Supported by the development of targeted analytical methodology (Arrivault et al., 2009 and 2015) and tailored labelling setups (Szecowka et al., 2013; Arrivault et al., 2017), these studies provided robust quantitative data on fluxes through central C metabolism and photosynthesis. The pioneering study of Szecowka and colleagues (Szecowka et al., 2013) has obtained the fluxes in canonical pathways of photosynthetic carbon metabolism in Arabidopsis rosette based on a combined 13CO2-labelling/GC- and LC-tandem MS/MS approach. Similar achievements have been made in Arabidopsis (under different conditions) (Ma et al., 2014), maize (Arrivault et al., 2017), tobacco (Chu et al., 2022), and Camelina sativa (Xu et al., 2021), but only partly in algae (Xiong et al., 2010; Wu et al., 2015) due to the experimentally challenging supply of rapidly labelled inorganic carbon pulses.
Using a tailor-made microfluidics labelling system to supply 13CO2 at steady state, we investigated in vivo labelling kinetics in intermediates of the Calvin Benson cycle and sugar, starch, organic acid, and amino acid synthesis pathways, and flux into protein and lipids, in several model and non-model green algae (Treves et al., 2022). Our system allows sampling at the 0–40 s pulse timescale in a highly reproducible manner, yielding high quality data. Furthermore, when combined with traditional labelling setups such as open systems with bubbling of 13CO2 for longer pulse times, our protocol, which is applicable to other unicellular photoautotrophic systems, largely improves the precision of flux estimates through the application of 13C-INST-MFA.
Materials and reagents
Inorganic 13C (Sigma-Aldrich, catalog number: 364592)
N2 gas (Oxygen and Argon Works Ltd.)
O2 gas (Oxygen and Argon Works Ltd.)
12CO2 gas (Oxygen and Argon Works Ltd.)
13CO2 (isotopic purity 99-atom percentage) gas (Sigma-Aldrich, catalog number: 364592-10L-EU)
Lime soda (Merck Millipore, catalog number: 1068395000)
Chlamydomonas reinhardtii strain CC-124 (UTEX Culture Collection of Algae)
Chlorella sorokiniana UTEX 1663 (UTEX Culture Collection of Algae)
Chlorella ohadii
HEPES (Sigma-Aldrich, catalog number: 54457)
Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: 655104)
Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C4901)
Ammonium chloride (NH4Cl) (Sigma-Aldrich, catalog number: 213330)
Magnesium sulphate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: 230391)
Dipotassium phosphate (K2HPO4) (Sigma-Aldrich, catalog number: P3786)
Monopotassium phosphate (KH2PO4) (Sigma-Aldrich, catalog number: PX1562)
Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884)
Boric acid (BO3H3) (Sigma-Aldrich, catalog number: B0394)
Zinc sulphate heptahydrate (ZnSO4·7H2O) (Sigma-Aldrich, catalog number: 221376)
Manganese(II) chloride tetrahydrate (MnCl2·4H2O) (Sigma-Aldrich, catalog number: 221279)
Iron(II) sulphate heptahydrate (FeSO4·7H2O) (Sigma-Aldrich, catalog number: 215422)
Cobalt(II) chloride hexahydrate (CoCl2·6H2O) (Sigma-Aldrich, catalog number: 255599)
Copper sulphate pentahydrate (CuSO4·5H2O) (Sigma-Aldrich, catalog number: 209198)
Ammonium molybdate tetrahydrate (Mn7O24(NH4)6·4H2) (Sigma-Aldrich, catalog number: 09878)
Methanol (Sigma-Aldrich, catalog number: 34860)
Chloroform (Sigma-Aldrich, catalog number: 650498)
tert-Butyl methyl ether (Sigma-Aldrich, catalog number: 650560)
Liquid nitrogen (N2)
Dry ice
Ethanol, technical
HP (HEPES-phosphate) medium (pH 7.2) (see Recipes)
Nutrient stock (see Recipes)
Phosphate buffer 1,000× (see Recipes)
Hutner’s solution (see Recipes)
MTBE (methyl tert-butyl ether) solution (see Recipes)
Equipment
Gas (mass-flow) controllers calibrated for N2, O2, and CO2 (Brooks Instruments, model: 5850S) and smart control software (Brooks Instruments)
20 mL syringes
Syringe pump (KF Technology, model: NE-1600 Multi-Channel Syringe Pump)
Transparent tubing (Vici Jour, catalog number: R-T-4007-M10)
Luer adapters (Upchurch Scientific, Union Polypropylene, UPP series)
Flangeless ferrule (Upchurch Scientific, catalog number: UPP-200N)
Union assembly (Upchurch Scientific, catalog number: UPP-360)
Luer adapter (Upchurch Scientific, catalog number: UPP-658)
Gas washing bottle (Robu Glasfilter-Geräte, catalog number: 41101)
Flat glass photobioreactors (PSI, model: FMT-150)
Cool-white LED array (PSI, model: 3500-D)
Centrifuge (Eppendorf, model: 5418 R)
LC-MS (Thermo Fisher Scientific, model: TSQ Quantum Ultra with Excalibur 2.07 SP1 and TSQ Quantum 1.4 software)
GC-MS (LECO Instruments GmbH, model: Pegasus III TOF-MS)
Gas chromatograph (Agilent Technologies, model: 6890N24)
Humidifier:
Laboratory bottles (DURAN, Sigma-Aldrich, catalog number: Z305197)
Head with filter-disc for GL 45 (ROBU Glasfilter-Geraete GmbH, catalog number: ISO 4793-80)
50 mL centrifuge tubes (Sarstedt, catalog number: 62.547.004)
Lyophilizer (Martin Christ, model: Alpha 2-4 LSCbasic)
Microfluidic mixer (Manufactured based on original design from Sivashankar et al., 2016)
Light meter (LI-COR, model: LI-250A)
Air pump (Sera precision, model: air 275 R plus, catalog number: 08814)
Autoclave (Witeg, model: WAC-60 230 V)
Filter 0.22 μm (Sigma-Aldrich, catalog number: SLMP025SS)
Water bath (GFL, model: 1002)
Oxygen sensor (Mettler Toledo, model: InPro 6800)
pH sensor (Mettler Toledo, model: InPro 3250)
Upright microscope (Nikon, model: Eclipse E200)
SpeedVac concentrator (Thermo Fisher Scientific, model: SPD210)
Gas analyser (LI-COR, model: LI850)
Procedure
Algal cultures
Grow algal cultures in flat glass bioreactor vessels, as specified in the instructions below. Further details associated with this technique are described in Treves et al. (2017).
Prepare HP medium (see Recipes) for the bioreactors.
Autoclave (121 °C, saturated steam, 30 min) each bioreactor cuvette (see https://photo-bio-reactors.com/products/photobioreactors/#details) with 1 L of HP medium and both oxygen and pH sensors connected.
On a sterile bench, connect 0.22 μm filters to all air inlets/outlets of the system and add 10 mL of axenic inoculum into the bioreactor medium, to reach an initial cell density of OD735nm = 0.02. The optical density of the culture is measured by the bioreactor following activation (see https://photo-bio-reactors.com/products/photobioreactors/#details).
Initiate the bioreactors at the optimal or target temperature defined for each strain and irradiate the cells at light regimes (in μmol photons m-2·s-1) optimised and designed for the study goals. For reliable estimation of in vivo fluxes, illumination levels used for labelling should be set to reflect the penetrating light levels within the bioreactor, as measured using an integrating sphere inside the running culture in the bioreactors.
Supply air for bubbling using an air-pump at approximately 1 L/min and monitor pH, dissolved oxygen concentration, and optical densities (OD) at 680 and 735 nm every 1–5 min, through integral bioreactor sensors (https://photo-bio-reactors.com/products/photobioreactors/#details).
Perform independent biological replicates of three separate bioreactor runs for each alga or condition.
Determine the weight of organic material within the samples through the measurement of ash-free dry weight per volume for each culture, as previously described in Klassen et al. (2015).
Note: Assess and validate the axenicity of the cultures with light microscopy and Luria-Bertani plating/incubation.
13CO2-labelling and sampling procedures
Two setups are used to provide inorganic 13C to algal liquid cultures (Figure 1).
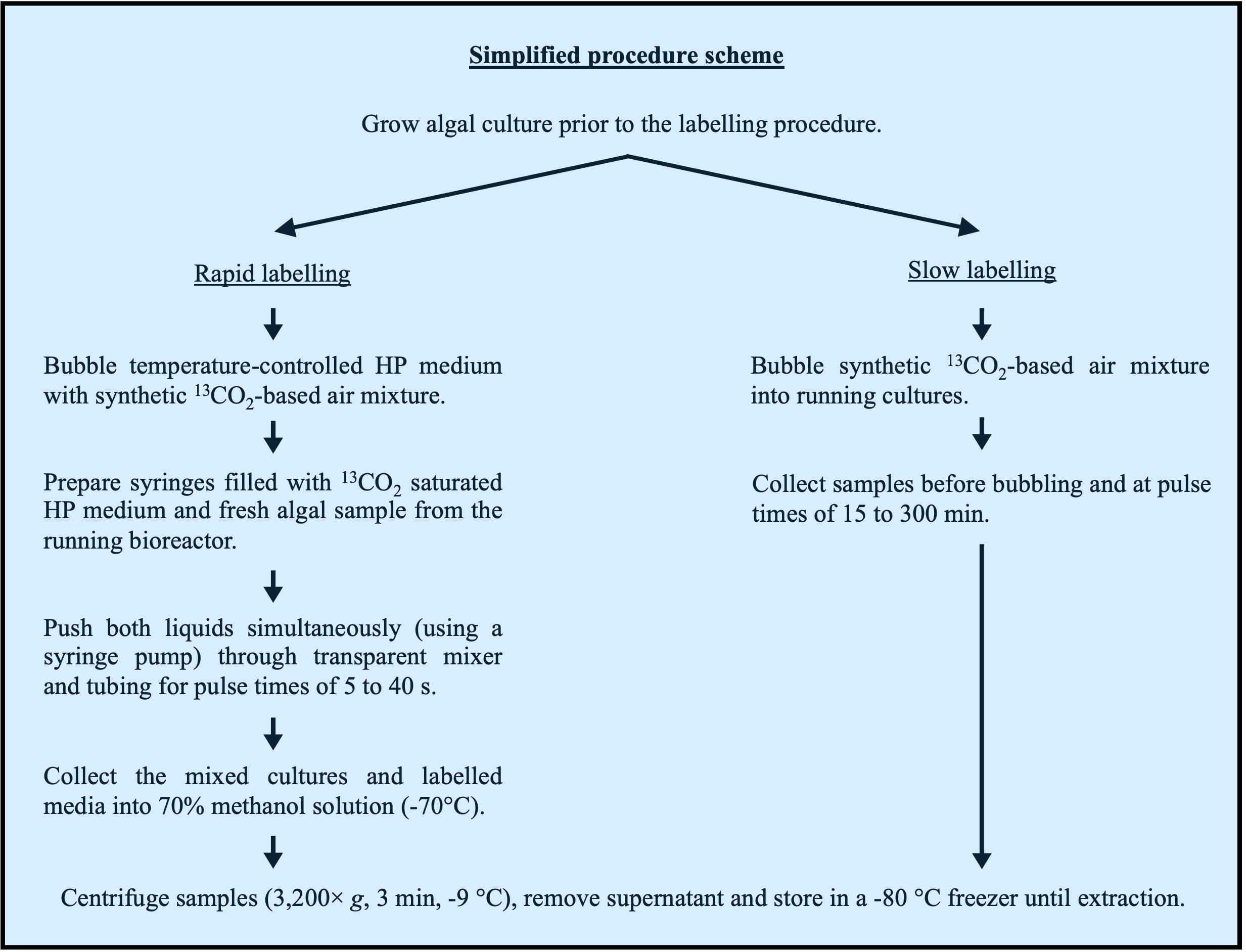
Figure 1. Simplified labelling procedure scheme. Flowchart presenting the main steps of rapid (left) and slow (right) labelling procedures.Very short pulses (up to 40 s) and rapid labelling (time points 5, 10, 20, and 40 s):
Fill a glass bottle with 400 mL of fresh HP medium (pH 7.2).
Place the glass bottle in a temperature-controlled water bath kept at the growth temperature of each algal culture, preferably in a separate room to avoid 13CO2 contamination in the bioreactors’ running cultures prior to mixing.
Bubble with the synthetic gas mixture (78% N2, 21% O2, and 400 ppm 13CO2) for 30 min to reach < 1% residual 12CO2 levels (see Figure 2B). Run exhaust flow through lime soda pellets via gas washing bottle to avoid 13CO2 cross contamination of running cultures.
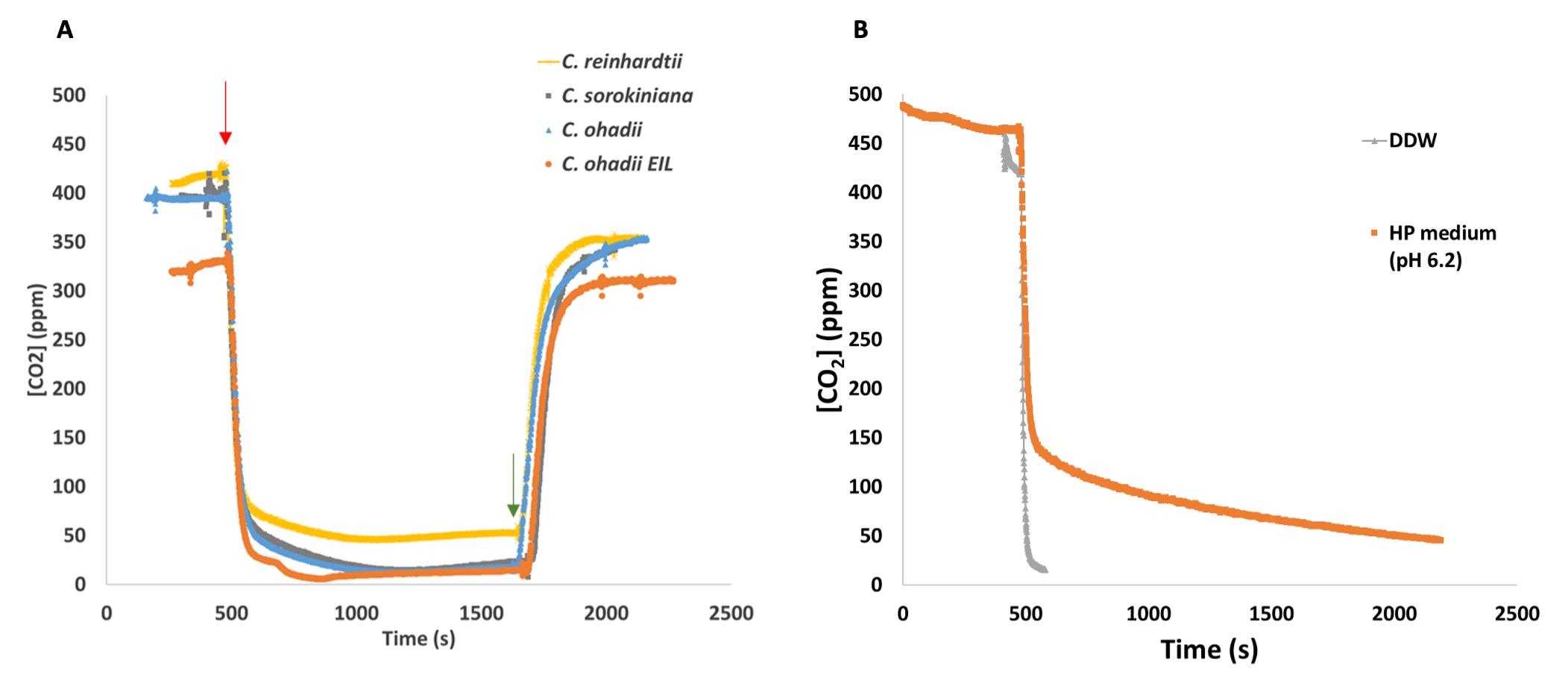
Figure 2. Kinetics of 13CO2 supply via bubbling (A) through algal cultures or (B) HP medium or double-distilled water (DDW). Monitoring of 12CO2 levels at the pulse, as measured on the exhaust of algal cultures/medium/DDW using a gas analyser. (A) Different algal cultures, grown to a density corresponding to T0 for each alga and condition, were bubbled with natural air. Thereafter, the natural air was rapidly changed (red arrow) to 13C-based synthetic labelled air mixture (see Procedure). Reciprocally, approximately 20 min after the first change, bubbling is rapidly replaced again for natural air (green arrow).Wash a 20 mL syringe three times and fill it with the bubbled solution (keep contact with air to a minimum).
Load the syringe to a syringe pump.
Connect the syringe with transparent tubing by screwing the luer to the screw tip of the syringe.
Turn on the light source above the syringe pump and bioreactor and adjust to illumination levels measured in step A4.
Connect transparent tubing (with length adjusted to pulse time, see Notes) between the mixer and the cooled 50 mL tubes (see Figure 3A).
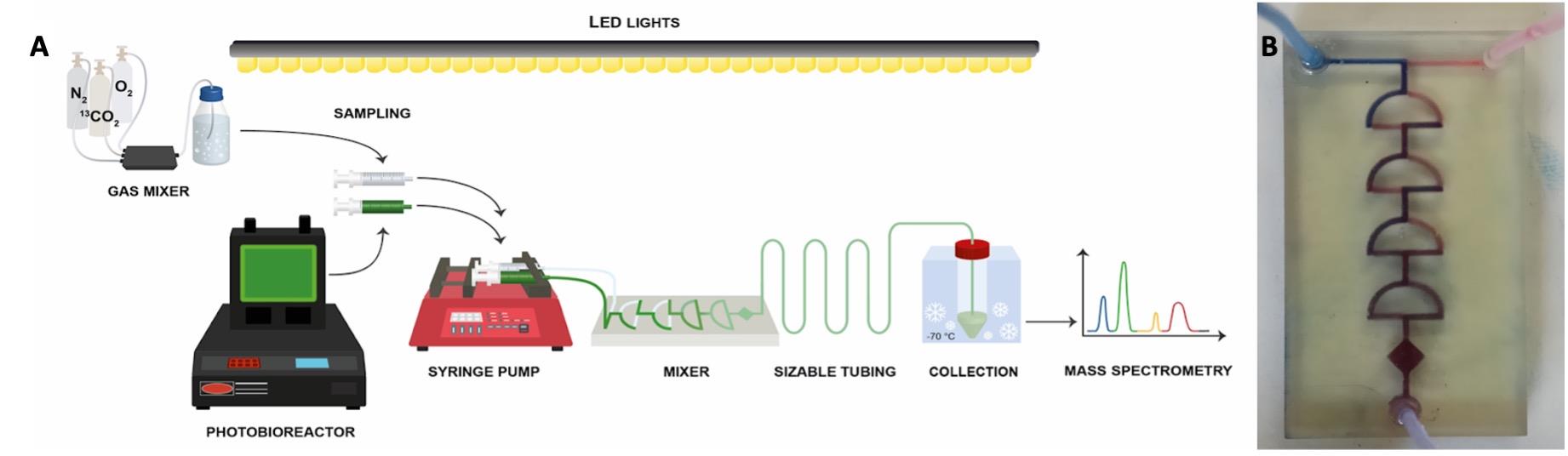
Figure 3. Rapid algal labelling system scheme. (A) Fresh medium bubbled with 13CO2-based synthetic air mixture (see Procedure) is mixed with fresh algal sample withdrawn from the bioreactor under controlled LED illumination. (B) Solutions are rapidly (< 1 s) mixed in a tailor-made transparent chip and injected through transparent tubing into 70% methanol solution kept at -70 °C. Pulse length (5–40 s) is controlled via tubing length.Withdraw a fresh algal sample into a 20 mL syringe using a transparent tube under the light. Load the syringe to the pump while avoiding any shading throughout the process.
Connect the syringe to transparent tubing by screwing the luer to the screw tip of the syringe (Figure 3A).
Operate the pump manually to push the liquids in both syringes and remove residual air from the tubes to the microfluidic mixer. Program the syringe pump to the required volume in each of the four tube lengths (see Section B, step 1h, above). Push the cultures and fresh media through the transparent mixer and tubing and collect the 1:1 mixture to a 70% methanol solution in 50 mL tubes cooled to -70 °C. Tubes are cooled in an ethanol bath precooled with dry ice (see scheme in Figure 3A). Inject the mixed sample into the cooled 70% methanol to reach a 1:2 ratio (e.g., 15 mL of sample into 30 mL of 70% methanol) (see Notes).
Keep the cooled 50 mL tubes at -70 °C until centrifugation (3,200× g, 3 min, -9 °C).
Discard the supernatant (see Notes) and freeze the pellet immediately in liquid nitrogen.
Repeat the same procedure for the non-labelled samples for each pulse time series replica by mixing the same algal culture with fresh HP medium pre-bubbled with ambient air at the same temperature as in Section B, step 1b.
Following rapid freezing, samples may be stored in a -80 °C freezer until extraction.
Resuspend the frozen pellets in ice-cold methanol/chloroform (5:1, v/v).
Perform four freeze-thaw cycles (see Notes) of resuspended cells and perform metabolite extraction following the methanol/chloroform procedure (see Mettler et al., 2014).
Notes:
Synthetic gas mixture is mixed using mass flow controllers as described previously (Szecowka et al., 2013).
Light is provided by an upper-positioned cool-white LED array through the entire route from sampling at the bioreactor to the quenching tube, with intensities corresponding to the penetrating light for each culture and avoiding shading.
Pulse duration is implemented by fitting the length of the transparent tubing downstream to the mixer to spray the culture/13CO2 solution mixture into the quenching tube at desired timings.
Metabolites level within the supernatant (see section B, step 1m) should be assessed by LC-MS/MS and should ideally be up to approximately 1% that of equivalent pellets.
Depending on the cell wall rigidity of the algal strains, the number of freeze-thaw cycles should be adjusted to the point where all chlorophyll migrates to the chloroform phase.
Microfluidic mixer (Figure 3B) may be produced by 3D printing, according to the detailed design provided in Sivashankar et al., 2016.
Long 13C incubation times (15–300 min) and slow labelling:
Introduce inorganic 13C via direct bubbling of the cultures with the gas mixture used for rapid labelling.
Bubble algal cultures with 13CO2 gas mixture (approximately 1 L/min) through a humidifier as done for the rapid labelling. Switch the gas mixture from natural air to the same synthetic 13C-labelled air mixture (T0). (Residual 12CO2 in the cultures should be below 2% within a few minutes of pulse. See Figure 2A). Run exhaust flow from labelled culture through lime soda pellets via gas washing bottle to avoid 13CO2 cross contamination of other running cultures.
Collect samples into 50 mL Falcon tubes before switching (T0) and at 15, 30, 60, 120, 180, and 300 min of bubbling with 13C synthetic gas mixture.
Centrifuge immediately (3,200× g, 3 min, -9 °C), remove supernatant, and freeze pellets in liquid nitrogen.
Resuspend frozen pellets in precooled (-20 °C) MTBE solution.
Perform four freeze-thaw cycles of resuspended pellets, followed by a two-phase MTBE extraction to analyse metabolites, protein, and starch, as described in Jüppner et al. (2017).
i. Dry the upper MTBE phase, containing lipids, in a SpeedVac concentrator and store at -80 °C to analyse lipid content and 13C labelling.
ii. Dry the lower phase, containing the polar and semipolar metabolites, in a SpeedVac concentrator and store at -80 °C for metabolite profiling.
iii. Store the solid pellets containing the precipitated protein and starch at -20 °C for further analysis of labelling into starch and protein as previously described (Bradford, 1976; Arrivault et al., 2009; Ishihara et al., 2015; Fernandez et al., 2017).
Notes:
Slow labelling is required, since longer incubation times would lead to Ci depletion in the closed microfluidic system.
Measured residual 12CO2 in the cultures reaches below 2% within a few minutes of pulse (Figure 2A) and H12CO3- is rapidly equilibrated by the cells as previously reported for low CO2-acclimated algae (Tchernov et al., 2003) and demonstrated here by the absence of a slow 12CO2 decay phase in the cultures (Figure 2A and 3B).
In step 2c, centrifugation is necessary as some metabolites can accumulate in the medium over time, thus leading to labelling patterns in the complete suspension not reflecting that in the cells.
Data analysis
Data analysis, peak annotation, and quality control (QC) procedures are thoroughly described in Arrivault et al. (2009) and Ma et al. (2014). Briefly, data analysis includes metabolite peak annotation and correction, followed by correction of raw data for the abundance of stable isotopes using the Corrector software tool (https://www.mpimp-golm.mpg.de/19405/Corrector_package_V1_91.zip), analysis of calibration curves for each metabolite and correction for signal decay along each run using periodic injection of standard mix solution; relative isotopomer abundance (m + n) for each metabolite is calculated as in Szecowka et al. (2013).
Recipes
All recipes regarding the preparation of the HP medium and associated stocks are available in Harris (1989).
HP medium, pH 7.2 (for 1 L)
HEPES 1.19 g
Nutrient stock 25 mL
Phosphate buffer 1,000× 1 mL
Hutner’s solution 1 mL
Make up to 1 L with DDW, set pH at 7.2 using NaOH, and autoclave.
Store in a glass bottle at room temperature (solution is not light sensitive).
Note: Following autoclave, add 25 mL of CaCl2 18 mM.
Nutrient stock (for 1 L)
NH4Cl 8.57 g
MgSO4·7H2O 4 g
Make up to 1 L with DDW and autoclave.
Store in a glass bottle at room temperature (solution is not light sensitive).
Phosphate buffer 1,000× (pH = 7) (for 1 L)
K2HPO4 106 g
KH2PO4 53 g
Make up to 1 L with DDW and autoclave.
Store in a glass bottle at room temperature (solution is not light sensitive).
Hutner’s solution, pH = 6.5–6.8 (for 1 L)
EDTA 50 g
Bo3H3 11.4 g
ZnSO4·7H2O 22 g
MnCl2·4H2O 5.06 g
FeSO4·7H2O 4.99 g
CoCl2·6H2O 1.61 g
CuSO4·5H2O 1.57 g
Mn7O24(NH4)·4H2O 1.1 g
Store in a glass bottle at 4 °C and wrap the glass bottle in aluminium foil (solution is light sensitive).
MTBE solution
Methanol/methyl tert‐butyl‐ether (1/3).
Store in a glass bottle at 4 °C (solution is not light sensitive).
Acknowledgments
We thank Dr. Stephanie Arrivault from MPI-MP for providing support for the optimization of algal harvest and extraction procedures through iterative LC-MS/MS measurements. We thank Prof. Dr. Mark Stitt for constructive discussions regarding experimental setup and controls design. We thank Dr. Hirofumi Ishihara for his support of integrating the labelling mixer with existing 13C-labelling gas mixer setup. This work was supported by grant 1697/22 from the Israeli Science Foundation and by the Alon fund from the Israel Council of Higher Education. This protocol was adapted from Treves et al. (2022).
Competing interests
The authors declare no competing interests.
References
- Abernathy, M. H., Yu, J., Ma, F., Liberton, M., Ungerer, J., Hollinshead, W. D., Gopalakrishnan, S., He, L., Maranas, C. D., Pakrasi, H. B., et al. (2017). Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol. Biofuels 10(1): e1186/s13068-017-0958-y.
- Adebiyi, A. O., Jazmin, L. J. and Young, J. D. (2015). 13C flux analysis of cyanobacterial metabolism. Photosynth. Res. 126(1): 19–32.
- Allen, D. K. and Young, J. D. (2020). Tracing metabolic flux through time and space with isotope labeling experiments. Curr. Opin. Biotechnol. 64: 92–100.
- Arrivault, S., Guenther, M., Fry, S. C., Fuenfgeld, M. M. F. F., Veyel, D., Mettler-Altmann, T., Stitt, M. and Lunn, J. E. (2015). Synthesis and Use of Stable-Isotope-Labeled Internal Standards for Quantification of Phosphorylated Metabolites by LC–MS/MS. Anal. Chem. 87(13): 6896–6904.
- Arrivault, S., Guenther, M., Ivakov, A., Feil, R., Vosloh, D., van Dongen, J. T., Sulpice, R. and Stitt, M. (2009). Use of reverse-phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J. 59(5): 826–839.
- Arrivault, S., Obata, T., Szecówka, M., Mengin, V., Guenther, M., Hoehne, M., Fernie, A. R. and Stitt, M. (2017). Metabolite pools and carbon flow during C4photosynthesis in maize:13CO2 labeling kinetics and cell type fractionation. J. Exp. Bot. 68(2): 283–298.
- Blank, L. M. and Sauer, U. (2004). TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiology 150(4): 1085–1093.
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254.
- Cheah, Y. E. and Young, J. D. (2018). Isotopically nonstationary metabolic flux analysis (INST-MFA): putting theory into practice. Curr. Opin. Biotechnol. 54: 80–87.
- Chu, K. L., Koley, S., Jenkins, L. M., Bailey, S. R., Kambhampati, S., Foley, K., Arp, J. J., Morley, S. A., Czymmek, K. J., Bates, P. D., et al. (2022). Metabolic flux analysis of the non-transitory starch tradeoff for lipid production in mature tobacco leaves. Metab. Eng. 69: 231–248.
- Fernandez, O., Ishihara, H., George, G. M., Mengin, V., Flis, A., Sumner, D., Arrivault, S., Feil, R., Lunn, J. E., Zeeman, S. C., et al. (2017). Leaf Starch Turnover Occurs in Long Days and in Falling Light at the End of the Day. Plant Physiol. 174(4): 2199–2212.
- Harris, E. (1989). The Chlamydomonas Sourcebook. A Comprehensive Guide to Biology and Laboratory Use. Academic Press.
- Heise, R., Arrivault, S., Szecowka, M., Tohge, T., Nunes-Nesi, A., Stitt, M., Nikoloski, Z. and Fernie, A. R. (2014). Flux profiling of photosynthetic carbon metabolism in intact plants. Nat. Protoc. 9(8): 1803–1824.
- Heise, R., Fernie, A. R., Stitt, M. and Nikoloski, Z. (2015). Pool size measurements facilitate the determination of fluxes at branching points in non-stationary metabolic flux analysis: the case of Arabidopsis thaliana. Front. Plant Sci. 6: e00386.
- Ishihara, H., Obata, T., Sulpice, R., Fernie, A. R. and Stitt, M. (2015). Quantifying Protein Synthesis and Degradation in Arabidopsis by Dynamic 13CO2 Labeling and Analysis of Enrichment in Individual Amino Acids in Their Free Pools and in Protein. Plant Physiol. 168(1): 74–93.
- Jazmin, L. J. and Young, J. D. (2013). Isotopically Nonstationary 13C Metabolic Flux Analysis. Methods Mol. Biol. : 367–390.
- Jüppner, J., Mubeen, U., Leisse, A., Caldana, C., Brust, H., Steup, M., Herrmann, M., Steinhauser, D. and Giavalisco, P. (2017). Dynamics of lipids and metabolites during the cell cycle ofChlamydomonas reinhardtii. Plant J. 92(2): 331–343.
- Klassen, V., Blifernez-Klassen, O., Hoekzema, Y., Mussgnug, J. H. and Kruse, O. (2015). A novel one-stage cultivation/fermentation strategy for improved biogas production with microalgal biomass. J. Biotechnol. 215: 44–51.
- Ma, F., Jazmin, L. J., Young, J. D. and Allen, D. K. (2014). Isotopically nonstationary 13C flux analysis of changes in Arabidopsis thaliana leaf metabolism due to high light acclimation. Proc. Natl. Acad. Sci. U. S. A. 111(47): 16967–16972.
- Mettler, T., Mühlhaus, T., Hemme, D., Schöttler, M. A., Rupprecht, J., Idoine, A., Veyel, D., Pal, S. K., Yaneva-Roder, L., Winck, F. V., et al. (2014). Systems Analysis of the Response of Photosynthesis, Metabolism, and Growth to an Increase in Irradiance in the Photosynthetic Model Organism Chlamydomonas reinhardtii . Plant Cell 26(6): 2310–2350.
- Niedenführ, S., Wiechert, W. and Nöh, K. (2015). How to measure metabolic fluxes: a taxonomic guide for 13 C fluxomics. Curr. Opin. Biotechnol. 34: 82–90.
- Sivashankar, S., Agambayev, S., Mashraei, Y., Li, E. Q., Thoroddsen, S. T. and Salama, K. N. (2016). A “twisted” microfluidic mixer suitable for a wide range of flow rate applications. Biomicrofluidics 10(3): 034120.
- Szecowka, M., Heise, R., Tohge, T., Nunes-Nesi, A., Vosloh, D., Huege, J., Feil, R., Lunn, J., Nikoloski, Z., Stitt, M., et al. (2013). Metabolic Fluxes in an Illuminated Arabidopsis Rosette. Plant Cell 25(2): 694–714.
- Tchernov, D., Silverman, J., Luz, B., Reinhold, L. and Kaplan, A. (2003). Massive light-dependent cycling of inorganic carbon between oxygenic photosynthetic microorganisms and their surroundings. Photosyn. Res. 77: 95–103.
- Treves, H., Küken, A., Arrivault, S., Ishihara, H., Hoppe, I., Erban, A., Höhne, M., Moraes, T. A., Kopka, J., Szymanski, J., et al. (2022). Carbon flux through photosynthesis and central carbon metabolism show distinct patterns between algae, C3 and C4 plants. Nat. Plants 8(1): 78–91.
- Treves, H., Murik, O., Kedem, I., Eisenstadt, D., Meir, S., Rogachev, I., Szymanski, J., Keren, N., Orf, I., Tiburcio, A. F., et al. (2017). Metabolic Flexibility Underpins Growth Capabilities of the Fastest Growing Alga. Curr. Biol. 27(16): 2559–2567.e3.
- Wieloch, T. (2021). The next phase in the development of 13C isotopically non-stationary metabolic flux analysis. J. Exp. Bot. 72(18): 6087–6090.
- Wu, C., Xiong, W., Dai, J. and Wu, Q. (2015). Genome-Based Metabolic Mapping and 13C Flux Analysis Reveal Systematic Properties of an Oleaginous Microalga Chlorella protothecoides . Plant Physiol. 167(2): 586–599.
- Xiong, W., Liu, L., Wu, C., Yang, C. and Wu, Q. (2010). 13C-Tracer and Gas Chromatography-Mass Spectrometry Analyses Reveal Metabolic Flux Distribution in the Oleaginous Microalga Chlorella protothecoides . Plant Physiol. 154(2): 1001–1011.
- Xu, Y., Fu, X., Sharkey, T. D., Shachar-Hill, Y. and Walker, a. B. J. (2021). The metabolic origins of non-photorespiratory CO2 release during photosynthesis: a metabolic flux analysis. Plant Physiol. 186(1): 297–314.
- Xu, Y., Wieloch, T., Kaste, J. A. M., Shachar-Hill, Y. and Sharkey, T. D. (2022). Reimport of carbon from cytosolic and vacuolar sugar pools into the Calvin–Benson cycle explains photosynthesis labeling anomalies. Proc. Natl. Acad. Sci. U. S. A. 119(11): e2121531119.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Geffen, O., Achaintre, D. and Treves, H. (2023). 13CO2-labelling and Sampling in Algae for Flux Analysis of Photosynthetic and Central Carbon Metabolism. Bio-protocol 13(17): e4808. DOI: 10.21769/BioProtoc.4808.
Category
Plant Science > Plant physiology > Photosynthesis
Plant Science > Plant metabolism > Carbohydrate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









