- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protocol for the High-quality Plasmid Isolation from Different Recalcitrant Bacterial Species: Agrobacterium spp., Rhizobium sp., and Bacillus thuringiensis
(*contributed equally to this work) Published: Vol 13, Iss 15, Aug 5, 2023 DOI: 10.21769/BioProtoc.4788 Views: 2493
Reviewed by: Hemant Kumar PrajapatiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Purification and Sequencing of DNA Guides from Prokaryotic Argonaute
Daan C. Swarts [...] John van der Oost
Nov 20, 2014 14656 Views
Evaluation of Plasmid Stability by Negative Selection in Gram-negative Bacteria
Damián Lobato Márquez and Laura Molina García
May 5, 2017 12865 Views

Conjugation Protocol Optimised for Roseburia inulinivorans and Eubacterium rectale
Paul O. Sheridan [...] Karen P. Scott
Apr 5, 2020 5493 Views
Abstract
High yield of good quality plasmid DNA from gram -ve bacteria (Agrobacterium tumefaciens, A. rhizogenes, and Rhizobium sp.) and gram +ve bacterium (Bacillus thuringiensis) is difficult. The widely used plasmid extraction kits for Escherichia coli yield a low quantity of poor-quality plasmid DNA from these species. We have optimized an in-house modification of the QIAprep Spin Miniprep kit protocol of Qiagen, consisting of two extraction steps. In the first, the centrifugation after adding neutralization buffer is followed by ethanol (absolute) precipitation of plasmid DNA. In the second extraction step, the precipitated DNA is dissolved in Tris-EDTA (TE) buffer, followed by an addition of 0.5 volumes of 5 M sodium chloride and 0.1 volumes of 20% (w/v) sodium dodecyl sulfate. After incubation at 65 °C for 15 min, the plasmid DNA is extracted with an equal volume of chloroform:isoamyl alcohol (CIA). RNase (20 mg/mL) is added to the upper phase retrieved after centrifugation and is incubated at 37 °C for 15 min. The extraction of the plasmid DNA with an equal volume of CIA is followed by centrifugation and is precipitated from the retrieved upper phase by adding an equal volume of absolute ethanol. The pellet obtained after centrifugation is washed twice with 70% (v/v) ethanol, air dried, dissolved in TE buffer, and quantified. This easy-to-perform protocol is free from phenol extraction, density gradient steps, and DNA binding columns, and yields high-quality plasmid DNA. The protocol opens an easy scale up to yield a large amount of high-quality plasmid DNA, useful for high-throughput downstream applications.
Key features
• The protocol is free from density gradient steps and use of phenol.
• The protocol is an extension of the QIAprep Spin Miniprep kit (Qiagen) and is applicable for plasmid DNA isolation from difficult-to-extract bacterial species.
• The protocol facilitates the direct transformation of the ligation product into Agrobacterium by skipping the step of E. coli transformation.
• The plasmids isolated are of sequencing grade and the method is useful for extracting plasmids for metagenomic studies.
Graphical overview
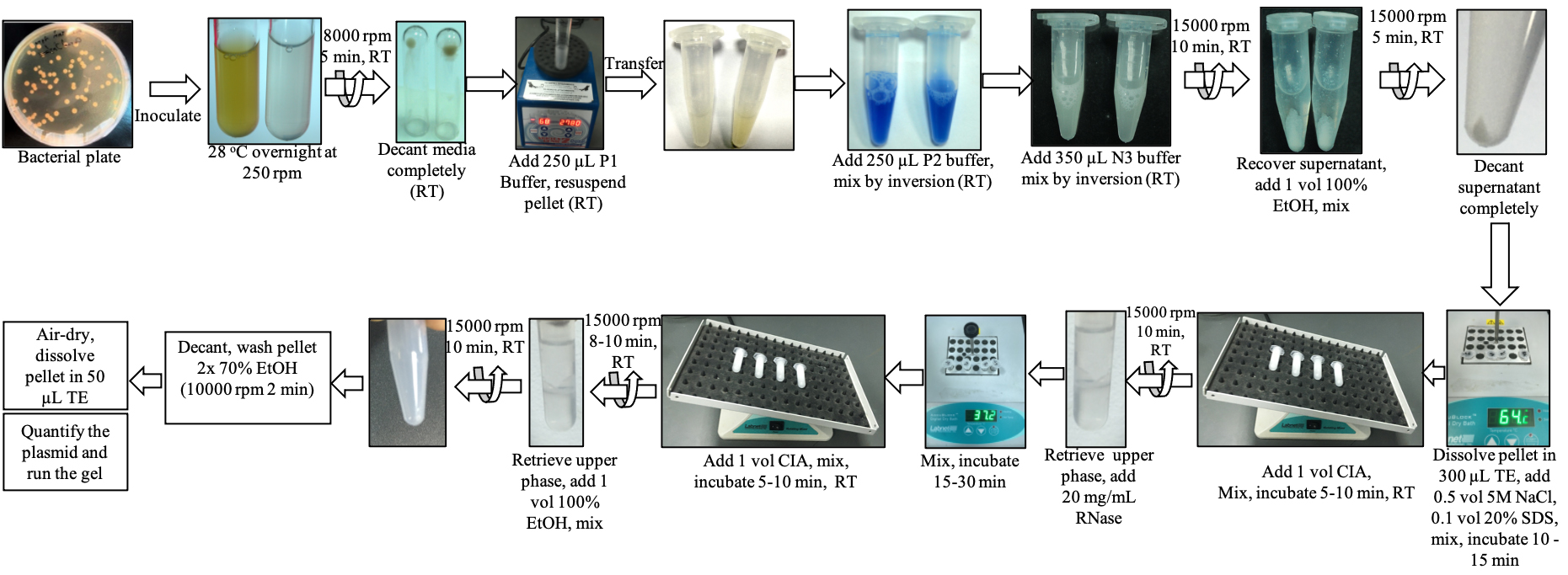
Overview of the plasmid isolation protocol (modified QIAprep Spin Miniprep kit) of the present study
Background
Plasmid isolation is an essential procedure in gene cloning, gene expression studies, sequencing, mutagenesis, and several downstream molecular processes. Easy-to-extract proprietary kits at different formats such as mini, midi, and maxi, and several published protocols are best suited for the plasmid extraction from widely used Escherichia coli strains. A simple and reliable method for isolating high-quality plasmid for downstream molecular applications from bacteria such as Agrobacterium tumefaciens, A. rhizogenes, Rhizobium sp., and Bacillus thuringiensis is lacking, due to the recalcitrance of the bacterial strains to cell lysis. Besides, lysozyme in the cell lysis solution is required to circumvent the non-lysis of the cell wall (Marmur, 1961). The Agrobacterium cells are not sensitive to the lysozyme-EDTA-detergent lysis procedures (Marmur, 1961; Clewell and Helinski, 1969), necessitating a relatively long treatment with proteolytic enzymes (Zaenen et al., 1974). Further, it is challenging to isolate Ti-plasmid free of chromosomal DNA (Ledeboer et al., 1976). In the case of agrobacteria, which is used to develop genetically modified and genome-edited plants, if there is no stock culture of E. coli with the desired plasmid, it necessitates retransformation of the plasmid into E. coli and subsequent extraction of plasmids for restriction digestion verification (Wise et al., 2006) and other downstream applications, e.g., biolistic transformation. There is no reliable protocol to extract high-quality plasmid DNA in large amounts from agrobacteria to accomplish downstream applications directly, i.e., without E. coli retransformation. The protocols described for the extraction of plasmid DNA from Agrobacterium, Rhizobium, and Bacillus thuringiensis are relatively lengthy and consist of sucrose gradients, CsCl-dye buoyant density gradients, or ethidium bromide treatment followed by phenol extraction (Ledeboer et al., 1976; Adachi and Iyer, 1980; Zhang and Kerr, 1993; Reyes-Ramirez and Ibarra, 2008). Our attempts to isolate plasmid from the gram -ve bacteria (Agrobacterium tumefaciens, A. rhizogenes, Rhizobium sp., and gram +ve bacterium (Bacillus thuringiensis) using kits (QIAprep Spin Miniprep kit from Qiagen and PureLink Quick Plasmid Miniprep kit from Invitrogen) and the user-modified protocol of QIAprep plasmid kit (Weber et al. 1998; https://www.qiagen.com/no/resources/resourcedetail?id=95083ccb-9583-489e-b215-99bd91c0604e&lang=en) did not yield satisfactory results. Extracting and purifying plasmid DNA from these strains and diverse other bacterial strains, recalcitrant to quality plasmid isolation, warrant a simple, short, and reliable protocol. We believe that the present study’s high-yielding, high-quality plasmid DNA protocol will be useful for other bacterial strains resistant to cell lysis, especially for low-copy number plasmid strains.
Materials and reagents
Biological materials
Agrobacterium tumefaciens strain EHA105, AGL1, GV3101, and LBA4404 [all containing binary plasmid pCAMBIA 1201 harboring gusA gene under the control of Cauliflower Mosaic Virus (CaMV) 35S promoter and bacterial selection marker chloramphenicol]
A. rhizogenes strains ATCC15834 and A4, containing binary plasmid harboring green fluorescent protein (GFP) gene under the control of 35S promoter and bacterial selection marker kanamycin
Rhizobium sp. (isolated from root nodules of desert tree legume, Prosopis cineraria)
Bacillus thuringiensis (received as a gift from a colleague, collected from the United Arab Emirates)
Reagents
Luria-Bertani (LB) broth (LB Miller Modification; PhytoTech Labs, catalog number: L475)
Tryptone (PhytoTech Labs, catalog number: T832)
Yeast extract (PhytoTech Labs, catalog number: Y892)
Sodium chloride (NaCl) (Sigma, catalog number: S1679)
Potassium dihydrogen phosphate dibasic (KH2PO4) (Sigma, catalog number: P3786)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma, catalog number: M2773)
Mannitol (Sigma, catalog number: M1902)
Bacto-agar (Himedia, catalog number: GRMO26)
Kanamycin sulfate (PhytoTech Labs, catalog number: K378)
Rifampicin (PhytoTech Labs, catalog number: R7382)
Formamide (Sigma, catalog number: 47671)
Chloramphenicol (PhytoTech Labs, catalog number: C1919)
QIAprep Spin Miniprep kit (Qiagen, catalog number: 27109)
PureLink Quick Plasmid Miniprep kit (Invitrogen, catalog number: K210011)
Resuspension solution P1 with RNase and Blue lysate (Qiagen, catalog number: 19051)
Resuspension buffer R3 (Invitrogen, catalog number: K2100-14)
Lysis solution P2 (Qiagen, catalog number: 19052)
Lysis buffer L7 (Invitrogen, catalog number: K2100-14)
Neutralizing solution N3 (Qiagen, catalog number: 19064)
Precipitation buffer N4 (Invitrogen, catalog number: K2100-14)
Sodium dodecyl sulfate (SDS) (Sigma, catalog number: L3771)
RNase (Thermo Scientific, catalog number: EN0531)
Ethanol absolute (Sigma, catalog number: NC1971050)
Disodium ethylene diamine tetraacetate (Na2EDTA·2H2O) (Sigma, catalog number: E6760)
Tris-base (Sigma, catalog number: T1503)
Hydrochloric acid (HCl) (Sigma, catalog number: 07102)
Sodium hydroxide (NaOH) (Sigma, catalog number: 06203)
Chloroform (Sigma-Aldrich, catalog number: C2432)
Glacial acetic acid (VWR, catalog number: VWRC20104.334)
Isoamyl alcohol (Sigma, catalog number: 19392)
Methanol ChromasolvTM (Sigma-Aldrich, catalog number: 34860-2.5L-R)
NcoI-HF (NEB, catalog number: R3193M), storage at -20 °C
BstEII-HF (NEB, catalog number: R3162M), storage at -20 °C
HotStar Taq DNA Polymerase PCR kit (Qiagen, catalog number: 203205)
PCR primers (Macrogen, South Korea)
Agarose D1 Low CE (Conda Lab, catalog number: 8010.00)
HydragreenTM Safe DNA Dye (20,000×) (ACTGene, catalog number: ACT-IDMG04)
6× DNA loading dye (NEB, catalog number: B7024A)
GeneRuler 1 kb Plus DNA ladder (Thermo Fisher Scientific, catalog number: SM1333)
Lambda DNA/EcoRI Plus HindIII marker (Thermo Fisher Scientific, catalog number: SM0191)
Solutions
1 N HCl
1 N NaOH
LB Agrobacterium medium (see Recipes)
Yeast Mannitol medium (see Recipes)
Tris-EDTA (TE; see Recipes)
Tris (see Recipes)
Na2EDTA (see Recipes)
Kanamycin (see Recipes)
Rifampicin (see Recipes)
Tris-acetate-EDTA (TAE, see Recipes)
Chloramphenicol (see Recipes)
5 M Sodium chloride (see Recipes)
20% (w/v) SDS (see Recipes)
Chloroform:isoamyl alcohol (CIA) (see Recipes)
Ethanol (70%, v/v) (see Recipes)
Media components (1 L) for the bacterial culture (see Recipes)
Antibiotics preparation (see Recipes)
Stock solutions (see Recipes)
Tris-EDTA (TE) buffer (100 mL) (see Recipes)
10× Tris-acetate-EDTA (TAE) (see Recipes)
Agarose gel (0.8%, w/v) preparation (see Recipes)
Recipes
Media components (1 L) for the bacterial culture
Components LB Agrobacterium (g/L) LB (g/L) YM (g/L) Tryptone 10 (5 g for LBA4404 strain) 10 Yeast extract 5 5 0.1 NaCl 5 10 0.2 KH2PO4 10 MgCl2·7H2O 0.2 Mannitol 10 Dissolve the components in 800 mL of MilliQ water and adjust to 1 L using MilliQ water. Transfer into a Duran bottle. Adjust the pH to 7.0 using 1 N HCl/1 N NaOH.
For solid media, add bacto-agar 15 g/L after adjusting the pH.
Autoclave at 121 °C (15 psi) for 15 min and store at room temperature (RT) for two months.Antibiotics preparation
Antibiotics Preparation and storage Working concentration Kanamycin Weigh 50 mg in a 1.5 mL Eppendorf, add 1 mL of sterile water, dissolve, and store at 4 °C for one week. 50 mg/L Chloramphenicol Weigh 25 mg in a 1.5 mL Eppendorf, add 1 mL of absolute ethanol, dissolve, and store at 4 °C for one week. 25 mg/L Rifampicin Weigh 20 mg in a 1.5 mL Eppendorf, add 1 mL of formamide*, dissolve and warp with aluminum foil, and store at 4 °C for one week. 20 mg/L *Methanol is an alternative solvent.
Stock solutions
Stock Preparation and storage 1 M Tris-HCl (pH 8.0) Dissolve 121.1 g of Tris-base in 800 mL of MilliQ water. Adjust the pH to 8.0 by adding concentrated HCl. Adjust to 1 L using MilliQ water. Transfer into a 1 L Duran bottle. Autoclave at 121 °C (15 psi) for 15 min. Store at RT for six months. 0.5 M EDTA (pH 8.0) Weigh 93 g of disodium EDTA·2H2O in ~400 mL of MilliQ. Stir vigorously on a magnetic stirrer with a stir bar. Adjust the pH to 8.0 with NaOH (~10 g of NaOH pellets). Adjust to 500 mL using MilliQ water. Transfer into a 500 mL Duran bottle. Autoclave at 121 °C (15 psi) for 15 min. Store at RT for six months. 5 M NaCl Dissolve 29.2 g of NaCl in 80 mL of MilliQ water and adjust to 100 mL using MilliQ water. Transfer into a 100 mL Duran bottle. Autoclave at 121 °C (15 psi) for 15 min. Store in a Duran bottle at RT for six months. SDS (20%, w/v) Weigh 20 g of SDS, dissolve it in MilliQ water, and adjust to 100 mL using MilliQ water. Transfer into a 100 mL Duran bottle. Autoclave at 121 °C (15 psi) for 15 min. Store at RT for six months. Caution: use face masks during preparation. Chloroform:isoamyl alcohol (24:1) (CIA) Add 2 mL of isoamyl alcohol in a 50 mL sterile Falcon tube and adjust to 50 mL by adding chloroform. Mix well and cover with aluminum foil. Store at RT for two weeks. Caution: prepare in a fume hood. 70% (v/v) ethanol Measure 35 mL of absolute ethanol in a 50 mL sterile Falcon tube, add 15 mL MilliQ water, and mix well. Store at RT for six months. Tris-EDTA (TE) buffer (100 mL)
Components Volume (mL) Final concentration Tris-HCl (pH 8.0) 1.0 10 mM Na2EDTA (pH 8.0) 0.2 1 mM MilliQ water 98.8 Autoclave in a 100 mL Duran bottle at 121 °C (15 psi) for 15 min and store at RT for six months. 10× Tris-acetate-EDTA (TAE)
Components Amount Concentration 1× 1× preparation Tris-base 48.5 g 40 mM Mix 10 mL of 10× with 90 mL Na2EDTA (pH 8.0) 20 mL 1 mM MilliQ water Glacial acetic acid 11.4 mL 20 mM Dissolve tris-base in approximately 800 mL of MilliQ water. Add acetic acid and EDTA. Adjust to 1 L. Transfer into 1 L Duran bottle. Store at RT for six months. Agarose gel (0.8%, w/v) preparation
Weigh 0.48 g of agarose and add into 60 mL of 1× TAE buffer in a 250 mL Erlenmeyer flask. Microwave for 2 min, cool to 50 °C, add 6 μL of Hydragreen, and cast in the gel tray with 15 wells combs.
Laboratory supplies
90 mm Petri dish (Sangon Biotech, catalog number: F611001)
Safe-lock tubes (2.0 and 1.5 mL) (Eppendorf, catalog number: 0030 120.094)
Pipettes (1,000, 200, 100, 20, and 10 μL) (Rainin, catalog number: L1000XLS+, L200XLS+, L100XLS+, L20XLS+, and L10XLS+, respectively)
Duran bottle [DURAN® ORIGINAL GL 45, catalog number: 10113399 (1,000 mL) and 10108298 (500 mL)]
Beakers (0.6 and 1 L) (Nalgene, catalog number: 1201-0600 and 1201-1000, respectively)
PYREX® 250 mL narrow mouth Erlenmeyer flask (Corning, catalog number: 4980-250)
Graduated cylinders (0.5 and 1 L) (Nalgene, catalog number: 3662-0500 and 3662-1000, respectively)
Pipette tips (1,000, 250, and 20 μL) (Rainin, catalog number: 30389294, 30389301, and 30389297, respectively)
Magnetic stirrer (Corning®, model: PC-620D, catalog number: 6796-620D)
Stir bars (PhytoTech Labs, catalog number: B011)
Tube racks (Globe Scientific, catalog number: 456350B)
Kimwipes (KIMTECH, catalog number: 34120)
14 mL sterile Falcon bacterial culture tubes (Falcon, catalog number: 352057)
Disposable gloves (Kimberly-Clark, catalog number: 52816)
Inoculation loops (VWR, catalog number: 10806-354)
Wide Mini-Sub Cell GT Horizontal Electrophoresis System, 15 cm × 7 cm tray, with gel caster and casting gates (Bio-Rad Laboratories, catalog number: 1704469)
Weigh dish (Thomas Scientific, catalog number: 3846D19)
Weigh paper (Thomas Scientific, catalog number: 9885G50)
Spatula (PhytoTech Labs, KS, USA, catalog number: S798)
Face mask (Euromed, China, catalog number: EM1834)
Disposable cuvettes (BrandTech, catalog number: 759075D)
Equipment
Autoclave (Tomy, model number: SX-700)
Weighing scale (Mettler Toledo®, model: MS603S/01)
Refrigerator (4 and -20 °C) (Evermed, model: BLCRF-370W)
pH meter (Thermo Fisher, model: Fisherbrand Accumet AB150, catalog number: 13636AB150B)
Laminar air flow cabinet (Esco, Horizontal Laminar Flow Cabinet, Gen 3, model: LHG-3AG-F8, catalog number: 2120387)
Bead sterilizer (PhytotTech Labs, model: ErgoSteri VT glass bead sterilizer, catalog number: S7520)
Shaker incubator (Eppendorf, model: New Brunswick Innova® 42, catalog number: M-1335-0004)
UV-Visible spectrophotometer (Thermo Scientific, model: Evolution 201, catalog number: 840-210600)
Centrifuge (Tomy, model: MX-307)
Vortexer (Scientific industries, model: Vortex-GenieTM 2T mixer, catalog number: SI-T266)
Heating block (Labnet International, AccuBlock, Digital dry bath, model: Labnet International D1100, catalog number: D1100-230V)
Fume hood (Esco, AscentTM Max Ductless Fume Hood, model: ADC-4BI, catalog number: 2040042)
Nutating Mixer (Labnet International, model: Labnet GyroMiniTM Nutating 3-D Mixer with dimpled mat, 120 V, catalog number: S0500)
NanoDropTM (Thermo Scientific, catalog number: ND2000C)
Microwave (Samsung, model: ME733K)
MilliQ water purification system (MilliQ, model: Elix integral 10)
Gel DocTM EZ imager (Bio-Rad Laboratories, catalog number: 170-8270)
PowerPacTM Basic (Bio-Rad Laboratories, catalog number: 164-5050)
Electrophoresis unit (Bio-Rad Laboratories, catalog number: 1704405)
Desktop (Dell, model: Optoplex 9020)
Thermal cycler (Bio-Rad Laboratories, model: T100)
Software and datasets
NanoDrop 2000 (version 1.6, free, Thermo Fisher Scientific)
Image Lab 6.0 (Bio-Rad Laboratories)
Insight2 (Thermo Fisher Scientific, catalog number: 837-002700)
Microsoft Excel for Mac (v16.69.1, 2022)
Procedure
Note: All steps are carried out at room temperature (RT) unless otherwise stated.
Streak the bacterial species: EHA105, AGL1, GV3101, and LBA4404 strains on solid LB Agrobacterium media supplemented with 20 mg/L rifampicin and 25 mg/L chloramphenicol; Bacillus thuringiensis on LB solid medium; A. rhizogenes strains ATCC15834 and A4 on YM solid medium with 50 mg/L kanamycin; and Rhizobium sp. on YM solid medium (see Recipes). Incubate for two days at 28 °C in an incubator.
Transfer bacterial culture from step 1 into 5 mL of liquid media in 14 mL bacterial culture tubes with appropriate antibiotics as above specified to each strain and incubate at 28 °C overnight on an orbital shaker at 250 rpm.
Note: Transfer a single colony for A. tumefaciens, Rhizobium, and Bacillus, and 2–3 colonies of A. rhizogenes from the solid medium to 5 mL liquid medium. Check the OD of the culture using the spectrophotometer; use culture media with no bacteria as blank. Cultures of A. tumefaciens with OD 0.5–0.7 and an OD of 0.8–1.0 for other strains are used for plasmid extraction. A 10 mL culture of slow-growing or low-copy number plasmid results in a high yield.
Centrifuge the cultures at 9,000× g (8,000 rpm) for 5 min.
Decant the supernatant. Remove the liquid media completely by pipetting out or by keeping the tube upside-down on a Kimwipe for 2–5 min. (Caution: do not disturb the pellet.)
Add 250 μL of resuspension solution (P1 buffer containing RNase and Blue lysate of QIAprep Spin Miniprep kit) into each tube and vortex to resuspend the cell pellet completely.
Transfer the resuspended bacterial pellet into 1.5 mL Eppendorf tubes using a pipette.
Add 250 μL of lysis solution (P2 buffer of QIAprep Spin Miniprep kit) and mix by gently inverting the tubes 5–6 times. The solution quickly turns blue and viscous, indicating bacterial lysis.
Add 350 μL of neutralizing solution (N3 buffer of QIAprep Spin Miniprep kit) and mix by inverting the tubes 5–6 times. The blue solution turns white, and the bacterial chromosomal DNA appears as a white precipitate.
Centrifuge the tubes at 20,000× g (15,000 rpm) for 10 min.
Carefully transfer the supernatant into new 1.5 mL Eppendorf tubes using a pipette. (Caution: do not disturb the white precipitate.)
Add an equal volume of absolute ethanol into each tube and mix by inverting the tubes a few times.
Centrifuge the tubes at 20,000× g (15,000 rpm) for 5 min.
Decant the supernatant and carefully remove the liquid completely using a pipette or by leaving the tube upside-down on a Kimwipe for 1–3 min.
Add 300 μL of TE buffer (pH 8.0) (see Recipes), 0.5 vol (150 μL) of 5 M NaCl, and 0.1 vol (45 μL) of 20% SDS. Mix by inversion and incubate at 65 °C in a heating block for at least 10 min.
Note: Make sure the DNA is completely dissolved in TE. If not, increase the volume of TE. If using 10% SDS, add 0.2 vol.
Add an equal volume (500 μL) of CIA (see Recipes) and keep shaking on a Nutating Mixer for at least 5 min.
Centrifuge at 20,000× g (15,000 rpm) for 7–10 min.
Carefully transfer the upper phase (~400 μL) into new 1.5 mL Eppendorf tubes, add 16 μL (20 mg/mL) of RNase, mix well by inversion, and incubate at 37 °C in a heated block for at least 15 min. Add an equal volume of CIA and keep shaking on a Nutating Mixer for at least 5 min.
Note: Take the heat block and keep it at RT after step 14 for a few minutes. Incubation of the vial after adding RNase in the block will also work.
Centrifuge at 20,000× g (15,000 rpm) for 7–10 min.
Carefully transfer the upper phase (300 μL) into new 1.5 mL Eppendorf tubes, add an equal volume of absolute ethanol, and mix well by inversion.
Centrifuge at 20,000× g (15,000 rpm) for 7–10 min.
Carefully decant or pipette out the liquid phase and add 500 μL of 70% ethanol. Tap the tube to release the pellet.
Centrifuge at 20,000× g (15,000 rpm) for 1–2 min and repeat step 21.
Centrifuge as above, pipette out the liquid completely, and air dry by keeping the tube open at RT for 2–5 min.
Add 50 μL (if the pellet is large, add more volume) of TE. Completely dissolve the pellet by finger tapping.
Measure the quantity of the plasmid DNA using NanoDrop (Figure 1A–1D; Table 1).
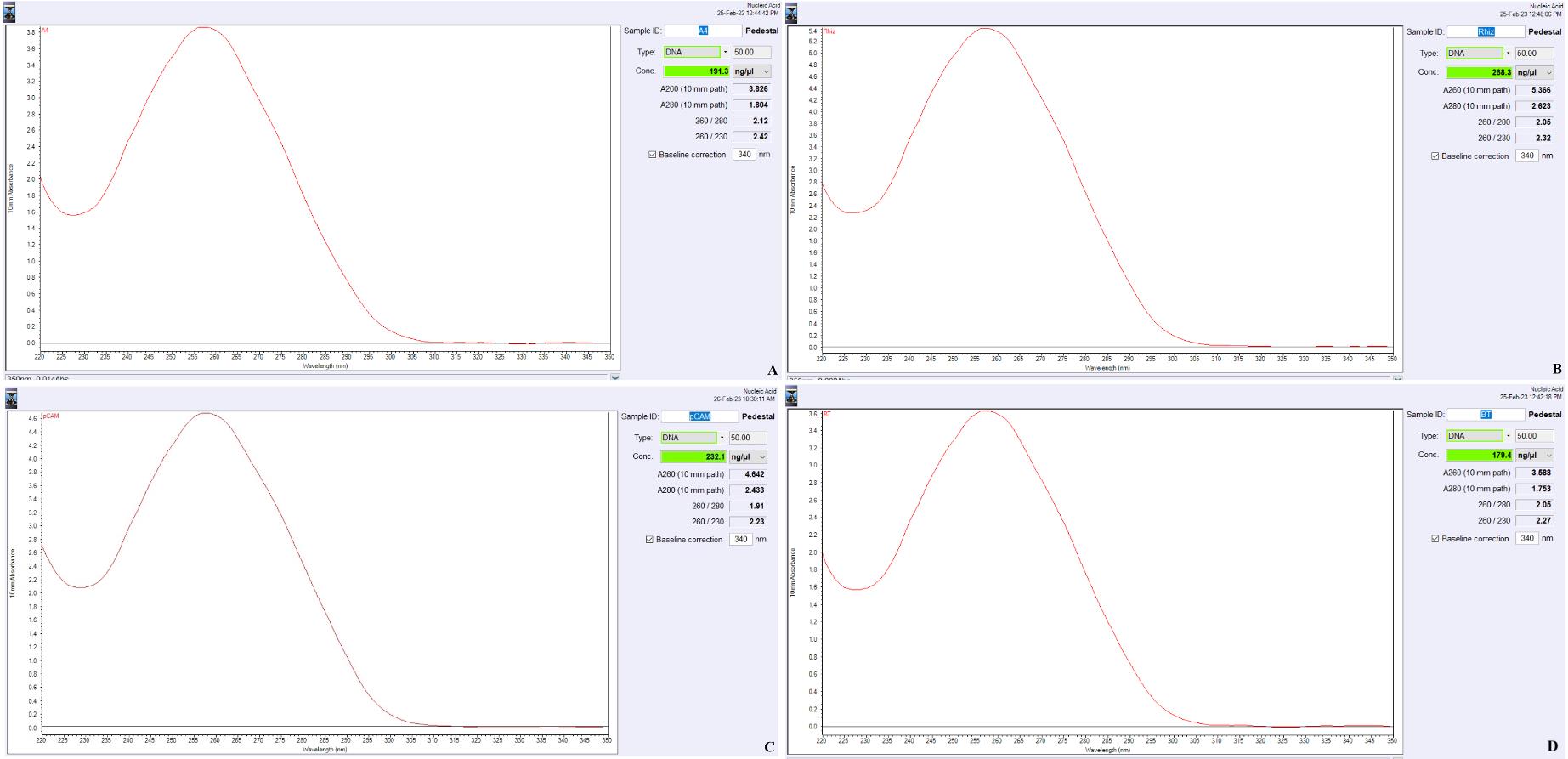
Figure 1. NanoDrop quantitation of the isolated plasmids. A. A4; B. Rhizobium; C. EHA105; and D. Bacillus thuringiensis. The x-axis represents the wavelength (nM), and the y-axis represents the 10 mm absorbance.Table 1. Purity and quantity of the plasmid isolated from different bacterial strains using Qiagen protocol for Agrobacterium (Weber et al., 1998) and the optimized protocol
Bacterial strain A260/280 ratio A260/230 ratio Yield (ng/μL) Control (Qiagen) Optimized protocol Control (Qiagen)
Optimized protocol Control
(Qiagen)
Optimized protocol A. tumefaciens EHA105 1.61 ± 0.03 1.91 ± 0.08 1.60 ± 0.04 2.10 ± 0.05 32 ± 1.8 232.4 ± 10.5 A. tumefaciens AGL1 1.40 ± 0.19 2.0 ± 0.05 1.52 ± 0.10 2.21 ± 0.1 24 ± 4.8 185 ± 8.6 A. tumefaciens GV3101 1.51 ± 0.07 1.93 ± 0.06 1.51 ± 0.07 2.09 ± 0.08 34 ± 1.4 212 ± 9.2 A. tumefaciens LBA4404 1.38 ± 0.14 2.05 ± 0.07 1.40 ± 0.10 1.98 ± 0.07 26 ± 3.4 176 ± 7.2 A. rhizogenes A4 1.49 ± 0.05 1.97 ± 0.08 1.49 ± 0.05 2.28 ± 0.1 22 ± 2.0 192.2 ± 9.8 A. rhizogenes ATCC15834 1.36 ± 0.11 1.99 ± 0.05 1.21 ± 0.08 2.15 ± 0.09 20 ± 3.1 183.1 ± 9.1 Rhizobium sp. 1.59 ± 0.03 1.89 ± 0.08 1.53 ± 0.04 2.11 ± 0.07 37 ± 4.3 372.0 ± 11.2 Bacillus thuringiensis 1.47 ± 0.12 1.91 ± 0.04 1.38 ± 0.11 2.10 ± 0.09 31 ± 3.5 211.7 ± 10.8 E. coli* 1.94 ± 0.06 - 2.09 ± 0.05 245 ± 8.1 - Data represent the mean ± SE of five replicates. The purity and quantity of plasmid isolated using PureLink Quick Plasmid Miniprep kit, which also served as the control, are not significantly different from that of the Qiagen protocol; hence, it is not shown. The same plasmid of that in A. tumefaciens strains in E. coli served as the positive control*.
Load 100 ng of plasmid DNA mixed with 6× loading dye (1× final) on a 0.8% agarose gel and run at 100 V for 25 min (Figure 2).
Document the gel using the Gel DocTM EZ Imager with the Image Lab 6.0 program.
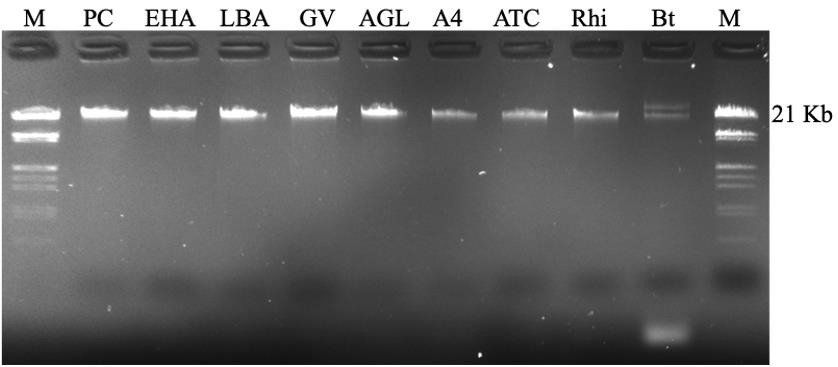
Figure 2. Gel electrophoresis of the isolated plasmids from different bacterial strains. EHA: EHA105, LBA: LBA4404, GV: GV3101, and AGL: AGL1 (all are A. tumefaciens); A4: A. rhizogenes A4; ATC: A. rhizogenes ATCC15834; Rhi: Rhizobium; Bt: Bacillus thuringiensis (shows two bands). PC: positive control of the same plasmid from E. coli using QIAprep Spin Miniprep kit; M: Lambda DNA/EcoRI Plus HindIII marker.
Data analysis
The quantity of the isolated plasmid DNA is determined using NanoDrop 2000 (Table 1; Figure 1A–1D). The data analyzed using Microsoft Excel represents the mean ± SE of five replicates (Table 1).
Validation of protocol
The protocol is repeated five times for each sample, once with a 10 mL culture of each bacterial strain, and control for each sample is carried out. Figure 2 shows the electrophoresis of the isolated plasmid DNA in an agarose (0.8%, w/v) gel in TAE containing Hydragreen (a noncarcinogenic nucleic acid stain), carried out in a Bio-Rad electrophoresis system, documented using the Gel DocTM EZ imager with the Image Lab 6.0 program. The quality of the plasmid DNA isolated in the present protocol is validated by gel electrophoresis (Figure 2) and restriction digestion (Figure 3): double digestion of 1 μg of plasmid DNA (pCAMBIA 1201) using NcoI-HF and BstEII-HF following the manufacturer’s double-digest protocol (NEB, USA). The digestion resulted in the gusA gene fragment of 2,060 bp (Figure 3). PCR is carried out using isolated plasmids with specific primers (Macrogen, Korea). Amplification of the gusA gene (Figure 4) of the extracted plasmid DNA from A. tumefaciens strains is carried out as per Kodackattumannil et al. (2023). PCR of the extracted Rhizobium plasmid with the primer pairs fD1 – rP2 for 16S rRNA (Weisburg et al., 1991) and R16-1 – R23-3R for ITS region (Figure 4) is performed as described by Kwon et al. (2005). Amplification of the GFP gene in the isolated plasmid DNA from A. rhizogenes strains (Figure 4) is accomplished according to Dutta et al. (2013). PCR of the Cry3a gene using the primers (forward: ATGAATCCGAACAATCGAAG and reverse: TTAATTCACTGGAATAAATTCAATTTTG) is carried out with an initial denaturation of 95 °C for 15 min, 30 cycles of denaturation at 94 °C for 30 s, annealing at 62 °C for 30 s, and extension at 72 °C for 2 min, and followed by a final extension at 72 °C for 10 min. In all cases, the PCR is carried out using HotStar Taq polymerase kit following the manual. The transient expression of GFP (plasmid isolated from ATCC15834) in indica rice IR5 callus (Figure 5), checked using particle bombardment (PDS-1000/Bio-Rad), validated the downstream application following the protocol of Sarangi et al. (2019).
Note: The isolated plasmid with the GFP gene (from ATCC15834) using the Qiagen protocol is not of good quality, and the quantity is very low (Table 1), which is insufficient to perform the particle bombardment.
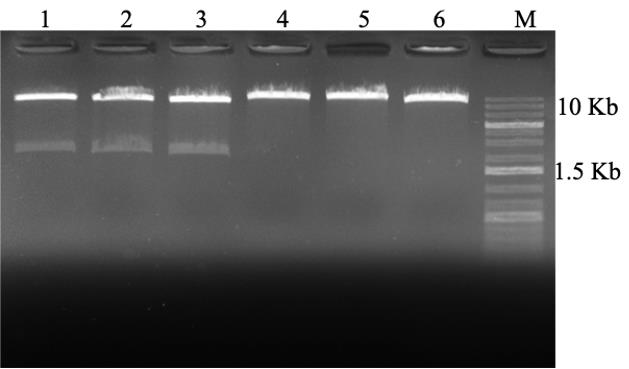
Figure 3. Restriction digestion of the plasmids isolated from A. tumefaciens strains EHA105, GV3101, and E. coli (positive control) containing the binary plasmid pCAMBIA 1201 harboring gusA (2,060 bp) gene under the control of 35S promoter (digestion of gusA gene using the restriction enzymes NcoI and BstEII (NEB). 1. EHA105; 2. GV3101; 3. E. coli; 4, 5, and 6 are undigested. M—1 Kb Plus DNA ladder.
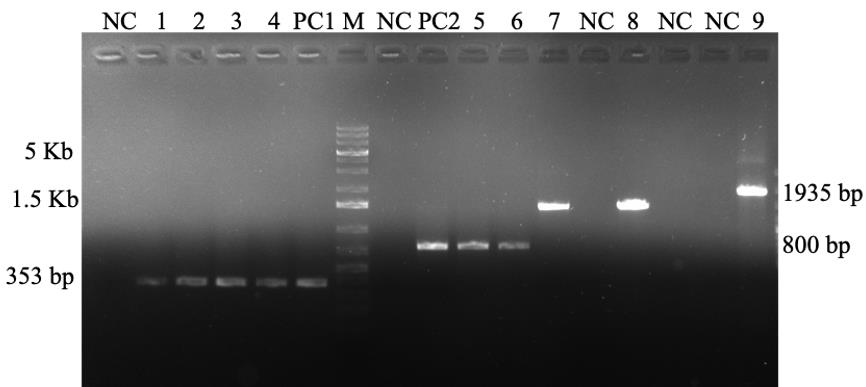
Figure 4. PCR analysis of the isolated plasmids from different bacterial strains. 1, 2, 3, and 4: gusA (353 bp), A. tumefaciens strain EHA105, GV3101, LBA4404, AGL1, and E. coli (PC1); 5 and 6: GFP (800 bp) from A. rhizogenes A4 and ATCC15834, respectively; 7 and 8: Rhizobium with R16-1–R23-3R, and fD1–rP2 primer pairs (1,500 bp); 9: amplification of cry3A gene (1,935 bp) from Bacillus thuringiensis. PC2: positive control of GFP; NC: negative control of the respective primers; M: 1 Kb Plus DNA ladder.
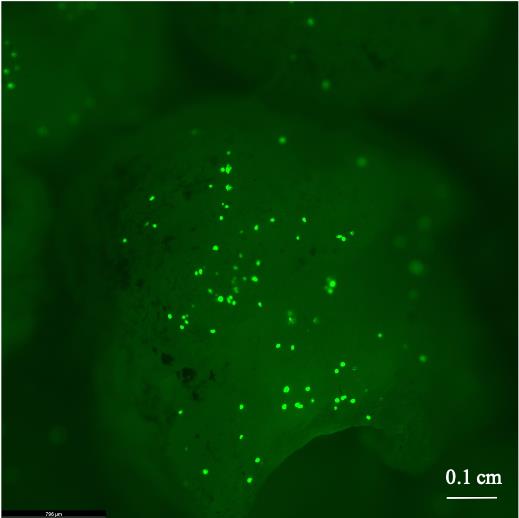
Figure 5. Green fluorescent protein expression on indica rice IR5 callus bombarded (using PDS-1000/Bio-Rad) as per the protocol of Sarangi et al. (2019) with the plasmid (harboring GFP gene under the control of 35S promoter) isolated from A. rhizogenes ATCC15834. Image is captured using Leica Thunder Model Organism Microscope with GFP filters (excitation 488 nm, emission 507 nm). Scale bar: 0.1 cm.
General notes and troubleshooting
Troubleshooting
| Problems | Troubleshooting |
| The quality of plasmid DNA is not good | a. Completely remove the bacterial culture medium before the addition of the P1 buffer. |
| b. Completely remove the ethanol before the addition of TE in the second extraction. | |
| SDS carry over | Do not use more than 0.1 vol of 20% SDS. |
| RNA contamination | Incubate at 37 °C for 30 min. |
Acknowledgments
We acknowledge The Presidential Court, the United Arab Emirates, for financial support.
Author contributions statement: MK designed the experiment. PK and SS optimized the protocol. SK and GL validated the protocol. MK and PK drafted the manuscript. KA supervised the project. All the authors reviewed the manuscript.
Competing interests
The authors declare no conflict of interest by any means.
Ethical considerations
The protocol has no animal or human subjects.
References
- Adachi, T. and Iyer, V. (1980). A procedure for the isolation and purification of plasmid DNA from Rhizobiummeliloti. Anal. Biochem. 101(2): 271–274.
- Clewell, D. B. and Helinski, D. R. (1969). Supercoiled circular DNA-protein complex in Escherichia coli: Purification and induced conversion to an open circular DNA from. Proc. Natl. Acad. Sci. U.S.A. 62(4): 1159–1166.
- Dutta, I., Kottackal, M., Tumimbang, E., Tajima, H., Zaid, A. and Blumwald, E. (2013). Sonication-assisted efficient Agrobacterium-mediated genetic transformation of the multipurpose woody desert shrub Leptadenia pyrotechnica. Plant Cell, Tissue Organ Cult. 112(3): 289–301.
- Kodackattumannil, P., Whitley, K., Sasi, S., Lekshmi, G., Krishnan, S., Al Senaani, S., Kottackal, M. and Amiri, K. M. A. (2023). Novel inducible promoter DREB1G cloned from date palm exhibits high fold expression over AtRD29 to drought and salinity stress. Plant Cell, Tissue Organ Cult.: e1007/s11240-023-02460-3.
- Kwon, S. W., Park, J. Y., Kim, J. S., Kang, J. W., Cho, Y. H., Lim, C. K., Parker, M. A. and Lee, G. B. (2005). Phylogenetic analysis of the genera Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium on the basis of 16S rRNA gene and internally transcribed spacer region sequences. Int. J. Syst. Evol. Microbiol. 55(1): 263–270.
- Ledeboer, A., Krol, A., Dons, J., Spier, F., Schilperoort, R., Zaenen, I., van Larebeke, N. and Schell, J. (1976). On the isolation of TI-plasmid from Agrobacterium tumefaciens. Nucleic Acids Res. 3(2): 449–464.
- Marmur, J. (1961). A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3(2): 208–218, IN1.
- Reyes-Ramírez, A. and Ibarra, J. E. (2008). Plasmid Patterns of Bacillus thuringiensis Type Strains. Appl. Environ. Microbiol. 74(1): 125–129.
- Sarangi, S., Mandal, C., Dutta, S., Mukherjee, P., Mondal, R., Kumar, S. J., Choudhury, P. R., Singh, V. P., Tripathi, D. K., Mandal, A. B., et al. (2019). Microprojectile based particle bombardment in development of transgenic indica rice involving AmSOD gene to impart tolerance to salinity. Plant Gene 19: 100183.
- Weber, S., Horn, R. and Friedt, W. (1998). Isolation of a low-copy plasmid from Agrobacterium using QIAprep technology. QIAGEN News, number: 5, 7.
- Weisburg, W. G., Barns, S. M., Pelletier, D. A. and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173(2): 697–703.
- Wise, A. A., Liu, Z. and Binns, A. N. (2006). Nucleic acid extraction Agrobacterium strains. In: Wang, K. (Ed.). Agrobacterium Protocols (pp. 67–76). Methods in Molecular Biology. Humana Press.
- Zaenen, I., van Larebeke, N., Teuchy, H., van Montagu, M. and Schell, J. (1974). Supercoiled circular DNA in crown-gall inducing Agrobacterium strains. J. Mol. Biol. 86(1): 109–127.
- Zhang, L. and Kerr, A. (1993). Rapid purification of Ti plasmids from Agrobacterium by ethidium bromide treatment and phenol extraction. Lett. Appl. Microbiol. 16(5): 265–268.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Kodackattumannil, P., Sasi, S., Krishnan, S., Lekshmi, G., Kottackal, M. and Amiri, K. M. A. (2023). Protocol for the High-quality Plasmid Isolation from Different Recalcitrant Bacterial Species: Agrobacterium spp., Rhizobium sp., and Bacillus thuringiensis. Bio-protocol 13(15): e4788. DOI: 10.21769/BioProtoc.4788.
Category
Microbiology > Microbial genetics > Plasmid
Molecular Biology > DNA > DNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









