- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Visualizing NBD-lipid Uptake in Mammalian Cells by Confocal Microscopy
Published: Vol 13, Iss 13, Jul 5, 2023 DOI: 10.21769/BioProtoc.4771 Views: 2390
Reviewed by: Jan HuebingerShalini Low-NamMario Ruiz

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Measurement of CMP-Sialic Acid Transporter Activity in Reconstituted Proteoliposomes
James Cahill [...] Matthew R. Whorton
Mar 20, 2020 4703 Views
Fluorescence Resonance Energy Transfer to Detect Plasma Membrane Perturbations in Giant Plasma Membrane Vesicles
Mathew Sebastiao [...] Steve Bourgault
Oct 5, 2023 2049 Views
PI(4,5)P2 Imaging Using a GFP Reporter in Living Cells
Mariam Alkandari [...] Mahtab Tavasoli
Jun 5, 2025 1877 Views
Abstract
Eukaryotic cells use a series of membrane transporters to control the movement of lipids across their plasma membrane. Several tools and techniques have been developed to analyze the activity of these transporters in the plasma membrane of mammalian cells. Among them, assays based on fluorescence microscopy in combination with fluorescent lipid probes are particularly suitable, allowing visualization of lipid internalization in living cells. Here, we provide a step-by-step protocol for mammalian cell culture, lipid probe preparation, cell labeling, and confocal imaging to monitor lipid internalization by lipid flippases at the plasma membrane based on lipid probes carrying a fluorophore at a short-chain fatty acid. The protocol allows studying a wide range of mammalian cell lines, to test the impact of gene knockouts on lipid internalization at the plasma membrane and changes in lipid uptake during cell differentiation.
Key features
•Visualization and quantification of lipid internalization by lipid flippases at the plasma membrane based on confocal microscopy.
•Assay is performed on living adherent mammalian cells in culture.
•The protocol can be easily modified to a wide variety of mammalian cell lines.
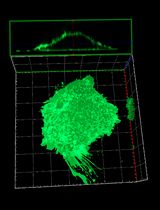
Graphical overview
Analysis of NBD-lipid uptake in adherent mammalian cells by confocal microscopy. Scale bar, 30 μm.
Background
Eukaryotic cells use a series of membrane transporters to control the movement of lipids across their plasma membrane. These transporters can be divided into ATP-dependent flippases and floppases—which catalyze the inward movement of phospholipids from the extracellular/luminal leaflet to the cytoplasmic leaflet and the outward movement of lipids, respectively—and ATP-independent scramblases (Holthuis and Levine, 2005; Contreras et al., 2010). Several tools and techniques have been developed to analyze the activity level of these transporters in the plasma membrane of mammalian cells. Among them, assays based on fluorescent lipid probes are particularly suitable, allowing visualization and quantification of lipid internalization by flippases in living cells. Lipid probes bearing a fluorophore such as nitrobenzoxadiazole (NBD) on a short-chain fatty acid at the sn-2 position, together with a long fatty acid residue at the sn-1 position, are most commonly used (Martin and Pagano, 1987; Koval and Pagano, 1991; Hoekstra and Kok, 1992; Rosenwald and Pagano, 1993) (Figure 1). Both BODIPY- and pyrene-labeled lipids have been used to track lipid trafficking in cells (Pagano et al., 1999; Somerharju, 2002), each having advantages and disadvantages (Table 1). Since the lipid polar head group stays unmodified, recognition as a substrate by ATP-dependent flippases and floppases is not affected (Theorin et al., 2019). At the same time, these molecules are less hydrophobic than their naturally occurring counterparts and readily insert into cell membranes when added to the medium. Transport of these short-chain lipid probes is usually monitored by extraction with bovine serum albumin (BSA) of the residual fraction of analogs not transported across the plasma membrane. As BSA extracts all analogs from the exoplasmic monolayer of the plasma membrane, the inaccessible fraction reflects analogs that have been internalized into cells. Alternatively, NBD-labeled lipids on the outer monolayer can be selectively destroyed with the water-soluble quencher dithionite (McIntyre and Sleight, 1991). However, because dithionite can leak through the membrane in some cells, conditions must be carefully adapted to the particular cell type (Pomorski et al., 1994). Furthermore, NBD-lipids are known to be actively metabolized by phospholipase activities. One frequent modification is their hydrolysis into lyso-derivates by phospholipase A2 activities, which results in the removal of the fatty acid attached to the sn-2 position. The liberated labeled C6 fatty acids are released into the medium, hampering the quantitative analysis of NBD-lipid internalization. Thus, the assay is typically performed in the presence of phospholipase inhibitors (Pomorski et al., 1996; Grifell-Junyent et al., 2022; Herrera et al., 2022).
The protocol presented here utilizes fluorescence microscopy to study NBD-lipid internalization in mammalian cells, exemplified on mouse skeletal muscle cell line (C2C12) cells (Grifell-Junyent et al., 2022), and has also been applied by us to fibroblasts (Pomorski et al., 1996). While NBD-lipids offer advantages (Table 1), including the ability to monitor the activity of lipid flippases at the plasma membrane of cells, it is important to note that these probes are less hydrophobic than naturally occurring lipids. This difference in hydrophobicity can affect the way they are transported within cells and may lead to differences in their intracellular trafficking compared to endogenous lipids. The protocol includes cell preparation, preparation of NBD-lipids, labeling of cells with NBD-lipids, fluorescence microscopy, and data analysis. The protocol can be easily adapted to parasites such as Toxoplasma and Leishmania (Weingärtner et al., 2011; Chen et al., 2021). It can also be applied to study: (i) the lipid uptake profile in mammalian cell lines; (ii) the impact of gene deletions and/or single mutations in lipid uptake at the plasma membrane; and (iii) the possible changes in lipid uptake during cell differentiation. For lipid uptake assays optimized for plants or based on flow cytometry, the reader is referred to previously published protocols (Jensen et al., 2016; López-Marqués and Günther Pomorski, 2021; Herrera et al., 2022).
Table 1. Main advantages and disadvantages of common fluorescently tail-labeled lipid probes
| Lipid probe | Advantages | Disadvantages | Reference |
|---|---|---|---|
| NBD-lipids | • Commercially available • Dithionite quenchable | • Low photostability • High environmental sensitivity | Martin and Pagano, 1987; Kobayashi and Arakawa, 1991; Pomorski et al., 1996) |
| BODIPY and TopFluor lipids | • High photostability • Low environmental sensitivity • Superior brightness • MαCD-mediated lipid exchange | • Availability restricted • Poorly extractable by BSA | (Pagano et al., 1999; Kay et al., 2012; Mioka et al., 2018; Segawa et al., 2021) |
Pyrene lipids | • Mimic natural lipids • Unique spectral features | • UV excitation • Low photostability • Availability restricted • Poorly extractable by BSA • High environmental sensitivity | Tanhuanpää et al., 2000; Somerharju, 2002 |
Abbreviation: MαCD, methyl-α-cyclodextrin
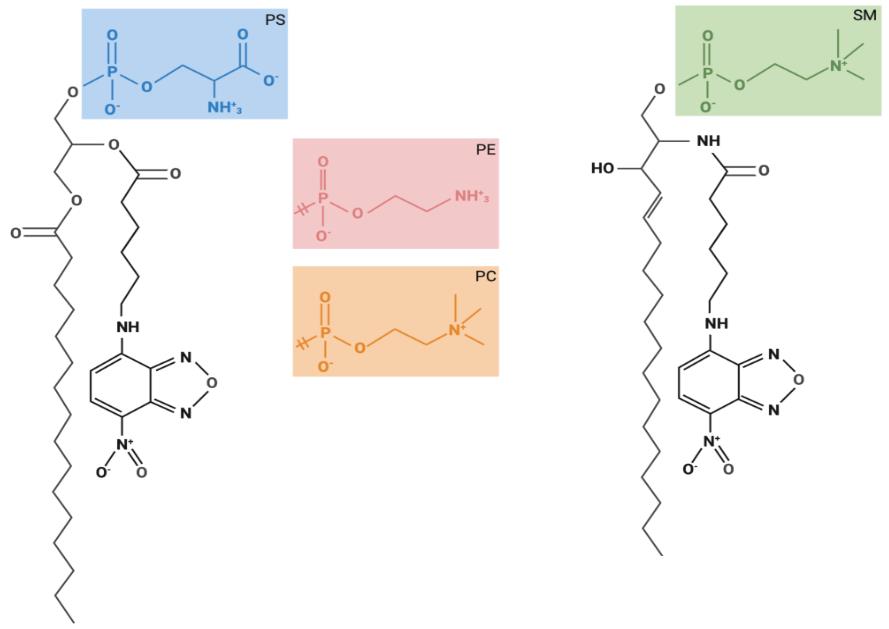
Figure 1. Chemical structures of fluorescent lipids used for lipid uptake assays. Glycerophospholipids are composed of two hydrophobic fatty acids and a hydrophilic head group, both combined with a glycerol backbone. Sphingolipids are composed of one hydrophobic fatty acid and a hydrophilic head group, both combined via a sphingosine backbone. The structures of the glycerophospholipid head groups corresponding to phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS), as well as for sphingomyelin (SM) are depicted. These fluorescently labeled lipids carry a nitrobenzoxadiazole (NBD) group at the sixth carbon in the short-chain fatty acid at the sn-2 position.
Materials and reagents
Mammalian cell culture
In this study, we used mouse myoblast cells (C2C12; cell number: ACC 565, DSMZ Braunschweig, Germany) that were cultured in growth medium (see Recipes). Optimal culture media and conditions may differ for other cell lines.
Dulbecco's Modified Eagle Medium, high glucose, without pyruvate (high-glucose DMEM) (e.g., Sigma-Aldrich, catalog number: D5796), store at 4 °C
Dulbecco's Modified Eagle Medium, low glucose, without pyruvate (low-glucose DMEM) (e.g., Sigma-Aldrich, catalog number: D6046), store at 4 °C
Ethanol absolute ≥ 99.8% (VWR, catalog number: 20821.321)
Fetal bovine serum, heat inactivated before use (FBS) (e.g., Capricorn Scientific, catalog number: FBS-11A), store at -20 °C
Hanks’ balanced salt solution, Ca2+ and Mg2+ free (HBSS) (e.g., Sigma-Aldrich, catalog number: H6648), store at 4 °C
Horse serum (HS) (e.g., Sigma-Aldrich, catalog number: H1138), store at -20 °C
35 mm polymer bottom dishes (e.g., Ibidi, catalog number: 81156)
1.5 mL microcentrifuge tubes (Sarstedt, catalog number: 72.690.001)
Penicillin-streptomycin, 100× solution (e.g., Sigma-Aldrich, catalog number: P4333), store at -20 °C
Pipette controller (e.g., accu-jet pro, Brand, catalog number: 263 00)
Polypropylene tubes of 15 mL capacity (e.g., Falcon tubes, Sarstedt, catalog numbers: 62.554.502 and 62.547.254)
Sterile serological pipettes (e.g., Serological pipettes of 5, 10, and 25 mL; Sarstedt, catalog numbers: 86.1253.001, 86.1254.001, and 86.1685.001)
Sterile culture vessels T-75 flasks (e.g., Sarstedt, catalog number: 83.3911)
Trypsin-EDTA solution (e.g., Sigma-Aldrich, catalog number: T3924), store at -20 °C
Trypan blue solution, 0.4% (Thermo Fischer Scientific, catalog number: 15250061)
Tyrode’s balanced salt solution (TBSS) (see Recipes), store at 4 °C
Preparation of NBD-lipids
Centrifuge glasses DURAN® with conical bottom, 12 mL (Carl Roth, catalog number: K211.1)
Chloroform 99%–99.4% ethanol-stabilized and certified for absence of phosgene and HCl (VWR, catalog number: 22711.290)
Methanol ≥ 99.8% (VWR, catalog number: 20847)
Glass vials (Rotilabo® screw neck ND8 vials, Brown glass, 1.5 mL, Carl Roth, Karlsruhe, Germany, catalog number: KE30.1) with screw caps (without borehole, without septum, PP, black, ND8, Carl Roth, Karlsruhe, Germany, catalog number: KE39.1) for lipid aliquoting
C16:0-C6:0 NBD-lipids purchased in chloroform including NBD-PC (Avanti Polar Lipids, catalog number: 810130), NBD-PE (Avanti Polar Lipids, catalog number: 810153), NBD-PS (Avanti Polar Lipids, catalog number: 810192), and NBD-SM (Avanti Polar Lipids, catalog number: 810218)
NBD-lipid uptake assay
Bovine serum albumin essentially fatty acid free (BSA) (Sigma-Aldrich, catalog number: A6003), store at 4 °C
Calcium chloride (CaCl2) (Grüssing, catalog number: 10043-52-4)
Dimethyl sulfoxide (DMSO) (Carl Roth, catalog number: 4720.4)
Glucose (Duchefa Biochemie, catalog number: G0802.5000)
4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid (HEPES) (Carl Roth, catalog number: 7365-45-9)
Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: 7791-18-6)
3-(4-octadecyl)benzoylacrylic acid, 4-(4-octadecylphenyl)-4-oxo-2-Butenoic acid (OBAA) (Sigma-Aldrich, catalog number: SML0075), store at -80 °C
Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626)
Note: OBAA is a potent inhibitor of phospholipase A2 (Eintracht et al., 1998). PMSF has been reported to inhibit phospholipases as well as some esterases (James, 1978; Gadella and Harrison, 2000; Estévez et al., 2012).
Potassium chloride (KCl) (Merck, catalog number: 7447-40-7)
Sodium chloride (NaCl) (Carl Roth, catalog number: 7647-14-5)
Sodium dihydrogen phosphate (NaH2PO4·2H2O) (VWR chemicals, catalog number: 28015.294)
Sodium hydroxide (NaOH) (Merck, catalog number: 30620-1KG-M)
Media and buffers
Growth medium (see Recipes)
Differentiation medium (see Recipes)
TBSS buffer (see Recipes)
NBD-lipid stocks (see Recipes)
PMSF stock of 200 mM (see Recipes)
OBAA stock of 5 mM (see Recipes)
BSA solution in TBSS (see Recipes)
Equipment
Autoclave sterilizer (e.g., Systec VX-65, Systec, Linden, Germany)
Biological safety cabinet certified for handling of biological materials (e.g., Herasafe KSP Class II Biological Safety Cabinets, Thermo Fisher Scientific)
Centrifuge with rotor for 15 mL polypropylene tubes (e.g., Eppendorf 5810 R; Wesseling, Germany)
Computer with monitor (e.g., DELL U2415)
Confocal laser scanning microscope (e.g., Leica TCS SP8)
Eppendorf Research® plus pipettes P2, P20, P200, P1000 (Eppendorf, catalog numbers: 3123000012, 3123000039, 3123000055, 3123000063)
Flow cabinet to work with organic solvents
Freezers -20 °C and -80 °C
Glass desiccator Boro 3.3 with socket in lid, 20 cm, including stopcock (BRAND, catalog number: 65238)
Hamilton 700 Series Syringes 10, 25, 100, and 1,000 μL (Hamilton Company, Nevada, USA)
Incubator with humidity and gas control to maintain 37 °C and 95% humidity in an atmosphere of 5% CO2 in air (e.g., Binder, Tuttlingen, Germany)
Incubator to maintain 20 °C (e.g., incubator with Peltier elements heating up and cooling down seamlessly in one system, Memmert, Schwabach, Germany)
Inverted phase contrast microscope equipped with a 10× objective (HI PLAN I 10×/0.22 PH1; Leica DMi1, Mannheim, Germany)
Neubauer counting chamber (improved dark lines, 0.1 mm) and cover glasses (20 × 26 × 0.4 mm)
Pipette tips 2, 10, 200, and 1,000 μL (Sarstedt, catalog numbers: 70.1130.212, 70.760.002, 70.3030.020, and 70.3050.020)
Refrigerator
Tubing (BRAND, catalog number: 143275)
Vortex mixer (e.g., Vortex Genie 2 Scientific Industries Inc., catalog number: SI-0236)
Vacuum Pump V-100 with Interface I-100 (Buchi, Switzerland, catalog numbers: 11593636 and 11593655D)
Water distillation system
Water bath (e.g., WPE45 Memmert, Schwabach, Germany) for mammalian cells and for NBD-lipid labeling (e.g., Julabo CORIO C-BT5, catalog number: 9011305)
Software
ImageJ (Wayne, Rasband, S., U. S. National Institutes of Health, Bethesda, Maryland, USA, https://imagej.nih.gov/ij/index.html, version v.153q)
Leica Application Suite AF (LAS X 3.5.7.23225, Leitz, Wetzlar, Germany)
Leica’s LAS X Office (LASX Office, ver. 1.4.4 26810, Leica, Wetzlar, Germany)
Procedure
The procedure below outlines four main steps: (A) preparation of mammalian cells, (B) cell differentiation, (C) cell counting, and (D) NBD-lipid uptake assay. This procedure was originally applied to study NBD-lipid internalization in mouse myoblast C2C12 cells (Grifell-Junyent et al., 2022). If this procedure is to be used with other cell types, adjustments might be required regarding the choice of cell culture medium, the requirement for coated surfaces, the cell density for microscopy, and possible detachment during incubation at low temperature (20 °C).
Preparation of mammalian cells
Grow adherent cells in sterile culture vessels (T-75 flask) in growth medium (see Recipes) in a tissue culture incubator (37 °C, 5% CO2, 95% humidity) until they reach ~60%–70% confluency.
Note: C2C12 cells will differentiate if grown too confluent and start to fuse. Differentiation and fusion are accompanied by transcriptional changes, which can lead to different results.
Aspirate and discard growth media with a sterile serological pipette.
Wash cells twice with 5 mL of HBSS (Ca2+ and Mg2+ free, pre-warmed at 37 °C) using a sterile serological pipette.
Add 1.5 mL of trypsin-EDTA solution (pre-warmed at 37 °C) using a sterile serological pipette and incubate the T-75 flasks in a tissue culture incubator (37 °C, 5% CO2, 95% humidity). Tilt the vessel back and forth a few times to make sure the thin layer of trypsin is evenly spread.
After 5 min, check for detachment by gently tilting the vessel and/or observing under an inverted microscope. If all cells have not detached in 5 min, incubate for an additional 1–2 min and check again. Continue to incubate and check as necessary, only until cells are no longer attached to the plate surface.
Note: Avoid prolonged incubation period with trypsin-EDTA solution.
Stop trypsinization by adding 7.5 mL of growth medium (pre-warmed at 37 °C, see Recipes) to the cell suspension.
Transfer the cell suspension into a 15 mL Falcon and set aside 100 μL in a 1.5 mL microcentrifuge tube for cell counting, e.g., using the hemocytometer (see section C).
Centrifuge cells in the 15 mL Falcon tube at 300× g for 5 min at room temperature and discard the supernatant to remove the trypsin-EDTA-containing medium from the cells.
Add 10 mL of fresh growth medium (pre-warmed at 37 °C, see Recipes) to the cell pellet and re-suspend completely by gently pipetting up and down using a serological pipette.
Note: Cells in suspension settle quickly. After counting, we recommend gently re-suspending the cell suspension approximately every 2–3 min when seeding multiple dishes.
After counting (see Section C), seed 1.5 × 104 cells per 35 mm polymer bottom dishes and add growth medium (pre-warmed at 37 °C, see Recipe 1) up to a final volume of 1 mL per dish.
Note: Prepare at least one dish per lipid to be analyzed plus one more for a negative control. We used a low cell number for seeding because C2C12 is a fast-growing cell line (doubling time: ~20 h) and this cell number guarantees that single cells are still present when the assay is performed the next day.
Keep the cells in a tissue culture incubator (37 °C, 5% CO2, 95% humidity) overnight.
Cell differentiation
To test the lipid uptake abilities and changes during differentiation of C2C12 cells, grow cells in 35 mm polymer bottom dishes in growth medium (pre-warmed at 37 °C, see Recipes) in a tissue culture incubator (37 °C, 5% CO2, 95% humidity) until they reach 100% confluency.
Note: To check for changes during differentiation and fusion, we analyze cells before they grow confluent (indicated as day -1), cells when they reach 100% confluency (indicated as day 0), and cells that were grown in differentiation medium for a specific number of days (Grifell-Junyent et al., 2022).
After reaching 100% confluency, aspirate and discard the growth medium.
Wash cells twice with 1 mL of HBSS (Ca2+ and Mg2+ free, pre-warmed at 37 °C).
Add 1 mL of differentiation medium (pre-warmed at 37 °C, see Recipes) per 35 mm polymer bottom dish and keep the cells in a humidified tissue culture incubator (37 °C, 5% CO2, 95% humidity).
Change the differentiation medium (pre-warmed at 37 °C, see Recipes) on a daily basis.
Note: We culture the cells up to seven days in differentiation medium.
Cell counting
The purpose of this step is to quantify the cell concentration to resuspend the cells at the appropriate concentration for the NBD-lipid uptake assay. We routinely use the trypan blue hemocytometer assay. Alternative cell counting methods, such as automatic cell counters, may be used.
Prepare the hemocytometer by cleaning the chambers and coverslip with ethanol. Dry the hemocytometer by using lint-free tissue. Place the glass coverslip over the counting chambers.
Note: The correct placement is indicated by the appearance of the Newton rings.
Add 100 μL of 0.4% trypan blue stock solution to 100 μL of cell suspension (step A6) to obtain a 1:1 dilution using a P200 Eppendorf pipette.
Load the hemocytometer with 10 μL of cell suspension per counting chamber with a P20 Eppendorf pipette and examine immediately under an inverted phase contrast microscope at low magnification (e.g., 5×–10× magnification).
Count the number of viable (seen as bright cells) and non-viable cells (stained blue) in the large outer quadrants.
Calculate the percentage of viable cells: % viable cells = [1.00 - (number of blue cells ÷ number of total cells)] × 100. Cell viability should be at least 95%.
Calculate the cell concentration based on the premise that each square accounts for a volume of 10-4 mL of cell suspension.
To obtain the total number of viable cells per milliliter of aliquot, multiply the total number of viable cells by 2 (the dilution factor for trypan blue) and the correction factor of 104 (volume of each square).
NBD-lipid uptake assay (see Figure 2)
The assay is typically performed at 20 °C to reduce endocytosis. For this, all solutions are pre-warmed at 20 °C before use and all incubation steps are performed in an incubator at 20 °C.
Prior to the start of the assay, prepare BSA solution in TBSS (see Recipes).
Prepare conical bottom glass tubes with NBD-lipid stocks (see Recipes) suspended in 10 μL of DMSO using a P20 Eppendorf pipette for each cell sample.
To prepare the cells for labeling, carefully aspirate and discard the growth medium using a P1000 Eppendorf pipette.
Wash cells twice with 1 mL of TBSS (pre-warmed at 20 °C, see Recipes) using a P1000 Eppendorf pipette.
After washing, add 1 mL of TBSS (pre-warmed at 20 °C, see Recipes) using a P1000 Eppendorf pipette.
To block the conversion of NBD-lipids by cellular phospholipases, add 5 μL of PMSF (see Recipes) and 1 μL of OBAA (see Recipes) to the cells using a P2 and P20 Eppendorf pipette to a final concentration of 1 mM and 5 μM, respectively,
Gently mix by tilting the dish and incubate for 10 min at 20 °C in an incubator.
To start labeling, remove 200 μL of TBSS from the cells in the dish, transfer into the conical bottom glass containing the DMSO NBD-lipid suspension (see step D2), mix well using a P200 Eppendorf pipette, and transfer the NBD-lipid suspension to the cells in the dish.
Gently mix by tilting the Petri dish and incubate for 60 min in a tissue culture incubator at 20 °C.
Note: If different NBD-lipids are to be tested, it is recommended to examine them one after the other rather than in parallel, as the time of microscopy would otherwise result in different timeframes of incubation.
After incubation, mount the dish on the microscope for imaging prior to BSA wash.
Note: At this point, cells can be tested for metabolic conversion of the NBD-lipids using lipid extraction and thin layer chromatography analysis, as previously described (Herrera et al., 2022).
For subsequent BSA back extraction, remove the dish from the microscope.
Remove TBSS and wash cells twice with 0.5 mL of BSA solution in TBSS (pre-warmed at 20 °C, see Recipes) for 1 min using a P1000 Eppendorf pipette.
Note: We routinely use a BSA concentration of 5% (w/v) for NBD-lipid extraction. However, the volume of BSA required for extraction, as well as the incubation time, may vary depending on cell type and lipid analog used in the assay (Fellmann et al., 2000). To determine the optimal conditions, label the cells at 4 °C and measure the cell-associated fluorescence after a different time of contact of the cells with BSA.
Add 1 mL of TBSS (pre-warmed at 20 °C, see Recipes) using a P1000 Eppendorf pipette and image on a confocal microscope.
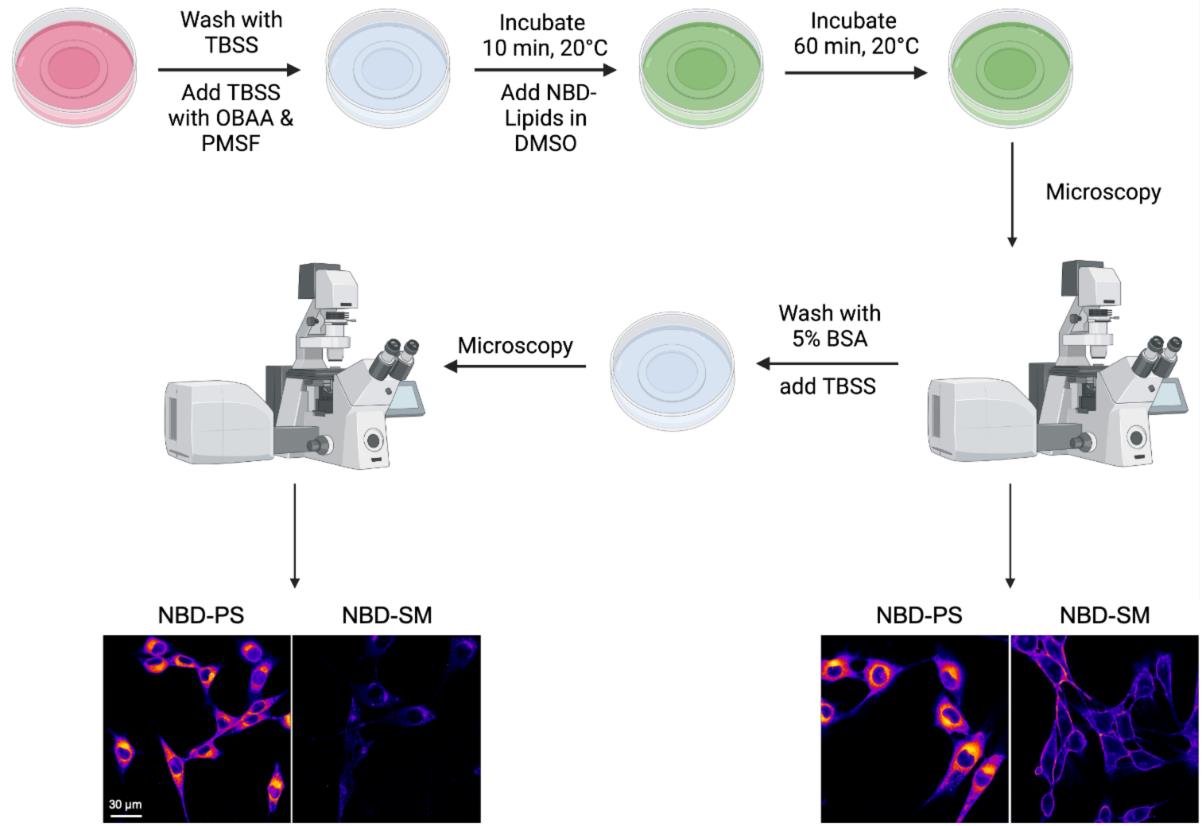
Figure 2. Schematic illustration of the nitrobenzoxadiazole (NBD)-lipid uptake assay via confocal microscopy. For labeling, cells pre-incubated with phospholipase inhibitors at 20 °C are supplemented with NBD-lipids and then incubated for up to 60 min to allow lipid internalization. After a TBSS wash, cells are analyzed using confocal fluorescence microscopy. Uptake of NBD-lipids results in intracellular labeling, while non-internalized NBD-lipids are found at the plasma membrane. Subsequently, back exchange with BSA is performed to remove non-internalized NBD-lipids at the plasma membrane, which allows visualizing only the internalized portion of the NBD-lipids.
Data analysis
Data acquisition
Images were acquired with a Leica TCS SP8 confocal laser scanning microscope (Leitz, Wetzlar, Germany) equipped with a 63×/1.20 water objective. For detailed imaging settings, see Table 1. All images were acquired at the same resolution, magnification, and orientation. This allows direct comparison of images and saves time when arranging figures.
Turn on the confocal laser scanning microscope and the laser, start the computer, and open the LAS X program.
The white light laser (WLL) intensity should be at 85%. Set the scan speed to 400 Hz. Situate the excitation to 488 nm with an intensity of 17.5%. Select a detector and switch it on. Configure the beam path manually to an emission of 497–600 nm with a gain of 20.3. For detailed imaging settings, see Table 2.
Note: The employed settings can be saved on the instrument to be reused in future experiments by selecting an image of a previous experiment with a right click and selecting Apply Image Settings.
Table 2. Parameters employed for confocal laser scanning microscopy analysis of the mammalian cell line C2C12 labeled with NBD-lipids. Settings employed for each parameter, including scanning, magnification, and information about the used objective, imaging settings, and intensities of the used lasers, excitation, and emission parameters. Note that these values might require adjustment depending on the cell line and/or microscope used.
Name Value Scan mode XYZ Logical size X/Y/Z 512/512/1 Physical length X/Y/Z 184.52 μm/184.52 μm/0 μm Scan direction X Unidirectional Scan speed 400 Hz Magnification 63 Objective name HC PL APO CS2 63×/1.20 Immersion Water Zoom 1 Frame average 1 Line average 6 WLL 85% Laser line 488 nm excitation 17.5% Emission 497 - 600 nm Gain 20.3 Transmission channel Gain 300 Detector for fluorescence HyD SMD 2 (HyD type detector) Detector for brightfield PMT Trans (PMT type detector) Choose the 63× magnification objective (water objective HC PL APO CS2 63×/1.20) and put a drop of water on the objective.
Note: The quality and resolution of an image is highly dependent on the choice of lens. One of the most important factors determining both sensitivity and resolution is the numerical aperture of the lens. To achieve the best possible image quality, it is strongly recommended to select a lens with a high numerical aperture. Care must be taken when using oil immersion objectives, as the 35 mm polymer bottom plates used in this protocol (see Materials) are only compatible with certain immersion oils.
Place the sample on the stage and search for the cells in brightfield mode.
To record the images, both brightfield and fluorescence signal, press the Live button. With the fluorescence signal on screen, search for representative areas and take several pictures.
Note: Exposure to laser light will cause bleaching. Therefore, it is recommended to adjust the focus in brightfield mode before switching to fluorescent mode and immediately acquire an image.
Save images on the computer as raw data.
Expected results
Depending on the head group and cell type, the lipid analog inserted into the outer plasma membrane leaflet can be internalized by spontaneous flip-flop, by protein-mediated translocation, or endocytosis. In C2C12 cells, for example, disappearance of NBD-phosphatidylserine (PS) from the cell surface is predominantly due to fast protein-mediated translocation across the plasma membrane (and endosomal membranes), resulting in a labeling of various intracellular membranes—including Golgi complex, ER, and endosomes. In contrast, NBD- sphingomyelin (SM) is internalized via endocytic vesicles resulting in the appearance of intracellular fluorescent spots (Figure 3).
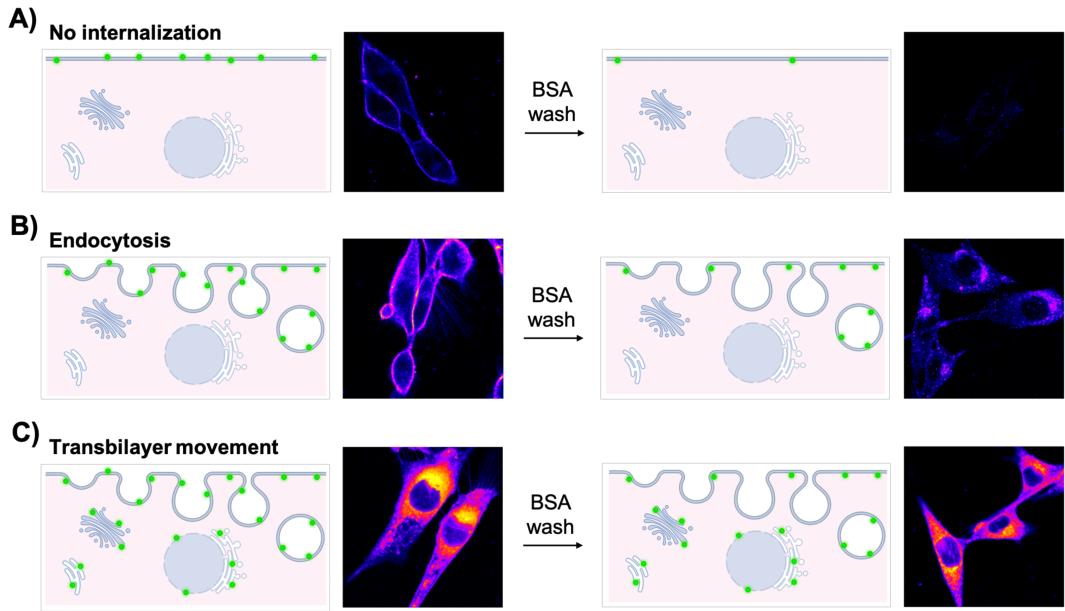
Figure 3. Interpretation of nitrobenzoxadiazole (NBD)-lipid labeling patterns.
Cells were incubated with NBD-lipids for 60 min and subjected to back exchange with BSA solution in TBSS. A) In the absence of endocytosis and active transport, NBD-lipids are internalized only when a spontaneous flip-flop occurs. Otherwise, NBD-lipids remain confined to the outer leaflet of the plasma membrane. Therefore, no residual fluorescence is found at the plasma membrane or in the cell interior after the BSA back extraction. B) If endocytosis takes place, fluorescence is mainly observed at the plasma membrane, but a weak signal is also visible inside the cell. After washing with BSA, low fluorescence can be detected inside the cell. C) For active lipid transport systems at the plasma membrane, a strong fluorescent signal is detectable throughout the cell before and after BSA back extraction.
Export the raw image data in a format compatible with ImageJ (e.g., tiff) from the imaging system.
Open the ImageJ software and import the images.
Click on Image in the upper operation row of the interface and change the image type to a 32bit grey scale.
Click again on Image and select the Fire Lookup Table (LUT) in the menu of different options for Lookup Tables.
Note: A LUT is a predefined spectrum of different shades of grey, each corresponding to a designated color. In case of the Fire LUT, a low intensity corresponds to dark grey and thus black and dark purple. A high intensity is converted to a light grey, which in turn corresponds to yellow and white. Therefore, a LUT is a tool to reflect differences in intensity.
Afterwards, select the Color menu under the Image operation and click on Edit LUT.
The LUT Editor will open, and the image colors will change to the corresponding colors of the Fire LUT. Press OK to set the change to the image.
Click on Image in the upper operation row of the interface and change the image type to RGB Color.
Save the images as a .tiff or .jpg file by selecting the File menu in the upper operation row of the interface and press Save As. Representative images are shown in Figure 4.
Note: All images to be compared should be imported and LUT-adjusted simultaneously.
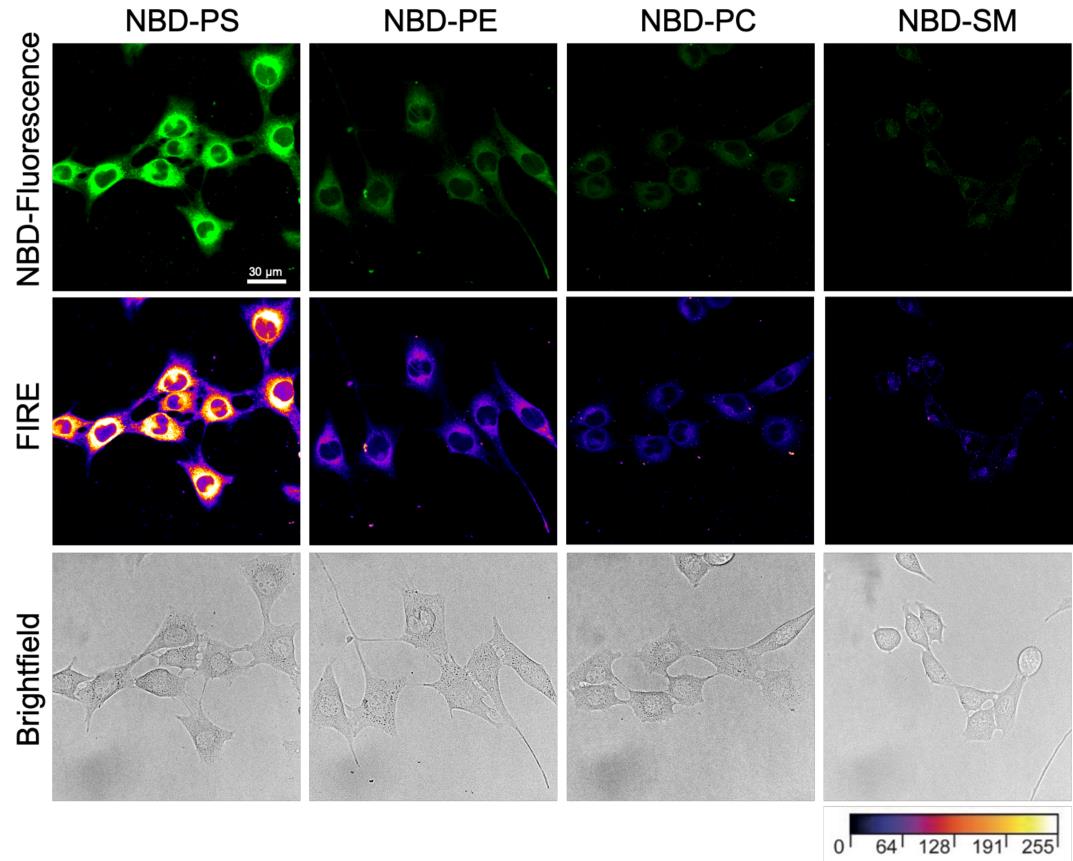
Figure 4. Representative confocal images of proliferating C2C12 wild-type cells. Cells were labeled with the indicated nitrobenzoxadiazole (NBD)-lipids for 60 min and subjected to back exchange with BSA solution in TBSS (pre-warmed at 20 °C, see Recipes). Images were taken by confocal microscopy and color-coded with the Fire Lookup Table to highlight intensity variations. Scale bar, 30 μm.
Quantification of lipid uptake
Import the raw data into Leica’s LAS X Office.
Click on the Quantify tab at the top of the screen and choose Histogram in the left selection area.
Note: To distinguish cells that are in close proximity and to improve the visibility of their outline for more precise gating, the intensity and contrast of the brightfield images can be adjusted.
Using the Polygon drawing tool located above the image, draw gates that encircle the cells closely, as shown in Figure 5.
Gate the cells based on the brightfield image so that gates will appear automatically and simultaneously at the same area on the fluorescence image.
Click on the Statistics bar, located just to the left of the images, to generate a table displaying values for the specific gate in both channels.
Deselect the brightfield channel and export the data table as .csv file format.
Note: The .csv file format is a standard file format that can be imported into other programs like Microsoft Excel or R for further data analysis. The analysis performed here was done on Microsoft Excel.
The exported data will give the pixel size of the drawn gate. Normalize the pixel size of each gate to 10,000 pixels.
Adjust the intensity sum accordingly with the ratio between the original pixel size and a pixel size of 10,000 for each gate.
Calculate the average intensity sum for a given lipid.
Calculate the standard deviation based on the adjusted intensity sum for each gate for a given lipid.
Plot data in a bar diagram (Figure 6).
Note: A negative control without NBD-lipid staining should be used to assess the level of background fluorescence in the sample. This is important because even in the absence of NBD-lipids, there may still be some level of background fluorescence present due to autofluorescence or other sources.
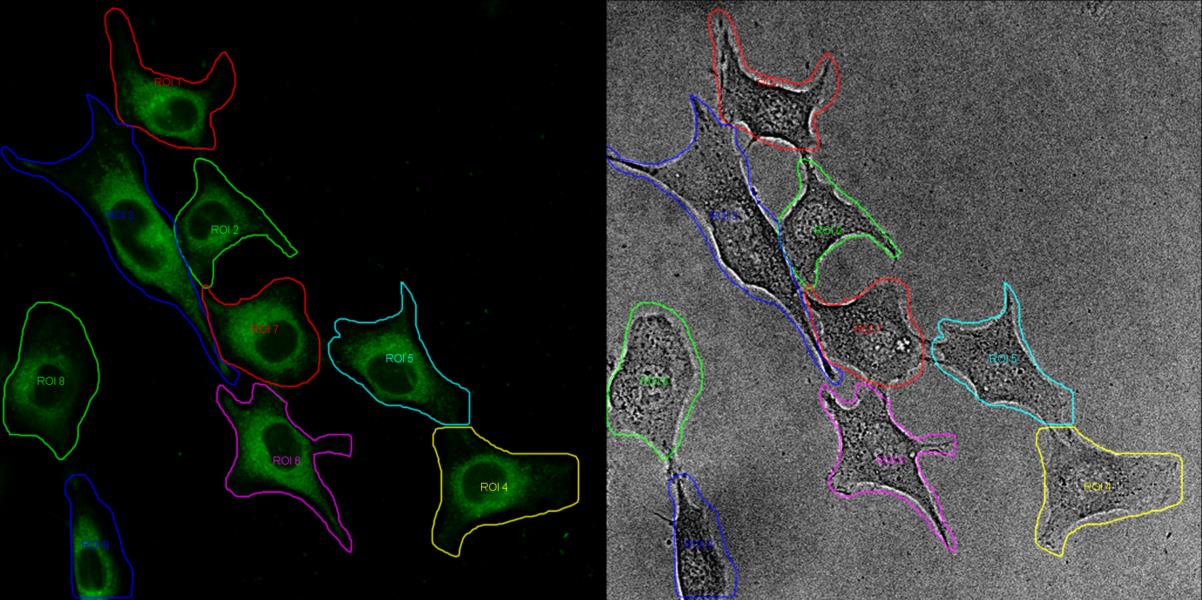
Figure 5. Gating of cells. The microscope images are processed using the Leica Application Suite LAS X software. Cells are selected based on the brightfield image and are then superimposed onto the fluorescence image by the software. The contrast and intensity of the brightfield image has been adjusted to facilitate the identification of the cells.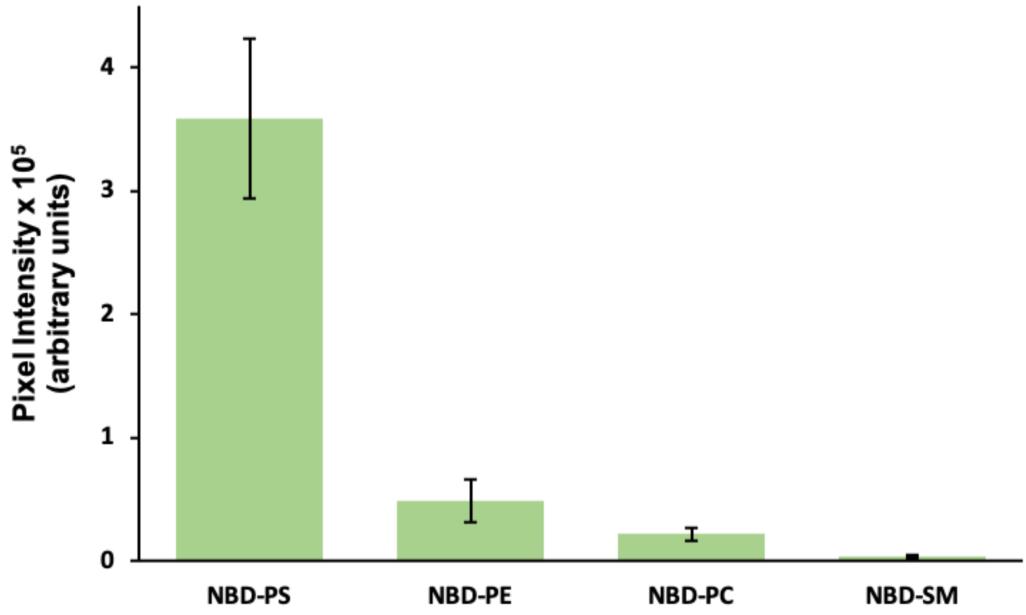
Figure 6. Average pixel intensity. Images of cells incubated with the indicated NBD-lipids were analyzed by single cell gating and pixel intensity analysis using Leica’s LAS X Office software. All gates are normalized to 10,000 pixels, with the intensity adjusted afterwards. Error bars represent standard deviation. In each case, 35 cells were analyzed from a representative experiment.
Recipes
Buffers were prepared using double-distilled water (ddH2O), which was obtained using an in-house water distillation system. Alternatively, all buffers are prepared using ultrapure water with purification sensitivity of 18 MΩ·cm-1 at 25 °C.
Growth medium
Open a 500 mL flask of high-glucose DMEM medium.
Add 100 mL of FBS.
Optional: Add 5 mL of 100× penicillin-streptomycin solution.
Prepare in sterile cabinet; store at 4 °C.
Differentiation medium
Open a 500 mL flask of low-glucose DMEM medium.
Add 10 mL of HS.
Optional: Add 5 mL of 100× penicillin-streptomycin solution.
Prepare in sterile cabinet; store at 4 °C.
TBSS buffer (1 L)
136 mM NaCl (7.94 g)
2.6 mM KCl (194 mg)
1.8 mM CaCl2 (200 mg)
1 mM MgCl2·6H2O (203 mg)
0.36 mM NaH2PO4·2H2O (56 mg)
5.56 mM glucose (1 g)
5 mM HEPES (1.2 g)
Adjust pH to 7.4 with 1 M NaOH. Complete volume to 1 L. Sterilize by filtering using a 0.22 μm filter. Store at 4 °C up to several months.
NBD-lipid stocks
All steps must be performed in glass tubes in order to prevent nonspecific binding of lipids.
Note: Chloroform and methanol are chemical hazards. Do not breathe gas/fumes/vapor/spray. Wear suitable protective clothing. Work in a fume hood.
Lipids are received suspended in chloroform, packaged in sealed glass ampoules; store at -20 °C until use.
To prepare lipid stocks, transfer a volume corresponding to 100 μg from the Avanti glass ampoule into glass screw neck vials. Evaporate the solvent from the glass vials in a desiccator at 250 mbar for 3 h with an additional 1 h incubation at 30 mbar. Close the vials with screw caps and parafilm and store at -20 °C until further use.
Remove desired lipid stocks from the freezer, place on ice, and dissolve in 1 mL of chloroform:methanol (1:1, vol) to a final lipid concentration of 100 μg/mL.
i. Use a glass syringe to transfer the desired volume of NBD-lipid stock solution into a 12 mm diameter glass tube. Typically, we use 5 nmol of NBD-lipid per dish.
ii. Dry the lipids under a 250 mbar vacuum overnight or under a gentle stream of nitrogen gas for 30 min to 1 h so that a dried lipid film is formed at the bottom of the tube.
iii. Resuspend the NBD-lipids in 10 μL of DMSO shortly before use. Add the DMSO in a circular fashion by placing the pipette tip onto the walls of the glass tube and on top of the dried lipid film. Slowly pipette up and down repeating the circular movement until all lipid film is resuspended.
Note: DMSO lipid suspensions are prone to precipitation, owing to their hygroscopic nature, and should be freshly prepared. DMSO is a chemical hazard. Wear suitable protective clothing.
PMSF stock of 200 mM
Weigh 34.838 mg with protective gear to avoid inhalation or contact with skin.
Dissolve in 1 mL of ethanol.
Store at -20 °C.
Note: PMSF is a chemical hazard, especially in its solid state, with high toxicity if swallowed and resulting in severe skin burns and eye damage upon contact. Wear suitable protective clothing and work in a fume hood when preparing the stock solution. Handle the stock solution carefully.
OBAA stock of 5 mM
Weigh 2.14 mg.
Add 1 mL of ethanol.
Store at -80 °C.
Note: Wear suitable protective clothing.
BSA solution (essentially fatty acid-free, 5% w/v) in TBSS
Weigh 200 mg of BSA in a 15 mL Falcon tube.
Add 4 mL of TBSS.
Incubate at 37 °C in water bath until dissolved.
Store in fridge until next day and use within one week; do not freeze the solution.
Note: This volume of 4 mL is enough to analyze the uptake of four NBD-lipids.
Acknowledgments
This protocol was adapted from our previous work (Grifell-Junyent et al., 2022; Herrera et al., 2022). The work was supported by the Lundbeckfonden (R221-2016-1005 to T.G.P.) and an instrument grant from the Deutsche Forschungsgemeinschaft (INST 213/886-1 FUGG to T.G.P.).
Competing interests
The authors declare that no competing interests exist.
References
- Chen, K., Günay-Esiyok, Ã., Klingeberg, M., Marquardt, S., Pomorski, T. G. and Gupta, N. (2021). Aminoglycerophospholipid flipping and P4-ATPases in Toxoplasma gondii. J. Biol. Chem. 296: 100315.
- Contreras, F. X., Sánchez-Magraner, L., Alonso, A. and Goñi, F. M. (2010). Transbilayer (flip-flop) lipid motion and lipid scrambling in membranes. FEBS Lett. 584(9): 1779–1786.
- Eintracht, J., Maathai, R., Mellors, A. and Ruben, L. (1998). Calcium Entry in Trypanosoma Brucei Is Regulated by Phospholipase A2 and Arachidonic Acid. Biochem. J. 336 (3): 659–666.
- Estévez, J., Barril, J. and Vilanova, E. (2012). Kinetics of inhibition of soluble peripheral nerve esterases by PMSF: a non-stable compound that potentiates the organophosphorus-induced delayed neurotoxicity. Arch. Toxicol. 86(5): 767–777.
- Fellmann, P., Herve, P., Pomorski, T., Muller, P., Geldwerth, D., Herrmann, A. and Devaux, P. F. (2000). Transmembrane movement of diether phospholipids in human erythrocytes and human fibroblasts.Biochemistry 39(17): 4994–5003.
- Gadella, B. M. and Harrison, R. A. P. (2000). The capacitating agent bicarbonate induces protein kinase A-dependent changes in phospholipid transbilayer behavior in the sperm plasma membrane. Development 127(11): 2407–2420.
- Grifell-Junyent, M., Baum, J. F., Välimets, S., Herrmann, A., Paulusma, C. C., López-Marqués, R. L. and Günther Pomorski, T. (2022). CDC50A is required for aminophospholipid transport and cell fusion in mouse C2C12 myoblasts. J. Cell Sci. 135(5): e258649.
- Herrera, S., Grifell-Junyent, M. and Pomorski, T. (2022). NBD-lipid Uptake Assay for Mammalian Cell Lines. Bio Protoc 12(4): e4330.
- Hoekstra, D. and Kok, J. W. (1992). Trafficking of glycosphingolipids in eukaryotic cells; sorting and recycling of lipids. Biochim. Biophys. Acta Biomembr. 1113: 277–294.
- Holthuis, J. C. M. and Levine, T. P. (2005). Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6(3): 209–220.
- James, G. T. (1978). Inactivation of the protease inhibitor phenylmethylsulfonyl fluoride in buffers. Anal. Biochem. 86(2): 574–579.
- Jensen, M. S., Costa, S. R., Theorin, L., Christensen, J. P., Pomorski, T. G. and López-Marqués, R. L. (2016). Application of image cytometry to characterize heterologous lipid flippases in yeast. Cytometry Part A 89(7): 673–680.
- Kay, J. G., Koivusalo, M., Ma, X., Wohland, T. and Grinstein, S. (2012). Phosphatidylserine dynamics in cellular membranes. Mol. Biol. Cell 23(11): 2198–2212.
- Kobayashi, T. and Arakawa, Y. (1991). Transport of exogenous fluorescent phosphatidylserine analogue to the Golgi apparatus in cultured fibroblasts.. J. Cell Biol. 113(2): 235–244.
- Koval, M. and Pagano, R. E. (1991). Intracellular transport and metabolism of sphingomyelin. Biochim. Biophys. Acta, Lipids Lipid Metab. 1082(2): 113–125.
- López-Marqués, R. and Günther Pomorski, T. (2021). Imaging of Lipid Uptake in Arabidopsis Seedlings Utilizing Fluorescent Lipids and Confocal Microscopy. Bio Protoc 11(22): e4228.
- Martin, O. C. and Pagano, R. E. (1987). Transbilayer movement of fluorescent analogs of phosphatidylserine and phosphatidylethanolamine at the plasma membrane of cultured cells. Evidence for a protein-mediated and ATP-dependent process(es). J. Biol. Chem. 262(12): 5890–5898.
- McIntyre, J. C. and Sleight, R. G. (1991). Fluorescence assay for phospholipid membrane asymmetry. Biochemistry 30(51): 11819–11827.
- Mioka, T., Fujimura-Kamada, K., Mizugaki, N., Kishimoto, T., Sano, T., Nunome, H., Williams, D. E., Andersen, R. J. and Tanaka, K. (2018). Phospholipid flippases and Sfk1p, a novel regulator of phospholipid asymmetry, contribute to low permeability of the plasma membrane. Mol. Biol. Cell 29(10): 1203–1218.
- Pagano, R. E., Watanabe, R., Wheatley, C. and Chen, C. S. (1999). Use of N-[5-(5,7-dimethyl boron dipyrromethene difluoride-sphingomyelin to study membrane traffic along the endocytic pathway. Chem. Phys. Lipids 102: 55–63.
- Pomorski, T., Herrmann, A., Zachowski, A., Devaux, P. F. and Müllery, P. (1994). Rapid determination of the transbilayer distribution of NBD-phospholipids in erythrocyte membranes with dithionite. Mol. Membr. Biol. 11(1): 39–44.
- Pomorski, T., Muller, P., Zimmermann, B., Burger, K., Devaux, P. and Herrmann, A. (1996). Transbilayer movement of fluorescent and spin-labeled phospholipids in the plasma membrane of human fibroblasts: a quantitative approach. J. Cell Sci. 109(3): 687–698.
- Rosenwald, A. and Pagano, R. (1993). Inhibition of glycoprotein traffic through the secretory pathway by ceramide.. J. Biol. Chem. 268(7): 4577–4579.
- Segawa, K., Kikuchi, A., Noji, T., Sugiura, Y., Hiraga, K., Suzuki, C., Haginoya, K., Kobayashi, Y., Matsunaga, M., Ochiai, Y., et al. (2021). A sublethal ATP11A mutation associated with neurological deterioration causes aberrant phosphatidylcholine flipping in plasma membranes. J. Clin. Invest. 131(18): e1172/jci148005.
- Somerharju, P. (2002). Pyrene-labeled lipids as tools in membrane biophysics and cell biology. Chem. Phys. Lipids 116: 57–74.
- Tanhuanpää, K., Virtanen, J. and Somerharju, P. (2000). Fluorescence imaging of pyrene-labeled lipids in living cells. Biochim. Biophys. Acta Mol. Cell Res. 1497(3): 308–320.
- Theorin, L., Faxén, K., Sørensen, D. M., Migotti, R., Dittmar, G., Schiller, J., Daleke, D. L., Palmgren, M., López-Marqués, R. L., Günther Pomorski, T., et al. (2019). The lipid head group is the key element for substrate recognition by the P4 ATPase ALA2: a phosphatidylserine flippase. Biochem. J. 476(5): 783–794.
- Weingärtner, A., dos Santos, M. G., Drobot, B. and Pomorski, T. G. (2011). Ca2+-activated transbilayer movement of plasma membrane phospholipids in Leishmania donovani during ionomycin or thapsigargin stimulation. Mol. Biochem. Parasitol. 179(2): 59–68.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Baum, J. F., Bredegaard, L., Herrera, S. A. and Pomorski, T. G. (2023). Visualizing NBD-lipid Uptake in Mammalian Cells by Confocal Microscopy. Bio-protocol 13(13): e4771. DOI: 10.21769/BioProtoc.4771.
Category
Cell Biology > Cell structure > Plasma membrane
Biochemistry > Lipid > Lipid transport
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link