- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
easyPACId, a Simple Method for Induced Production, Isolation, Identification, and Testing of Natural Products from Proteobacteria
(*contributed equally to this work) Published: Vol 13, Iss 13, Jul 5, 2023 DOI: 10.21769/BioProtoc.4709 Views: 1620
Reviewed by: Dennis J NürnbergSelcuk HazirAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Biosynthesis and Genetic Encoding of Non-hydrolyzable Phosphoserine into Recombinant Proteins in Escherichia coli
Philip Zhu [...] Richard B. Cooley
Nov 5, 2023 2493 Views
Mobilization of Plasmids from Bacteria into Diatoms by Conjugation Technique
Federico Berdun [...] Eduardo Zabaleta
Mar 5, 2024 1908 Views

Artificial Metalloenzymes in Artificial Sanctuaries Through Liquid–Liquid Phase Separation
Kaixin Wang [...] Tong Wu
Oct 5, 2025 1536 Views
Abstract
The easyPACId (easy Promoter Activation and Compound Identification) approach is focused on the targeted activation of natural product biosynthetic gene clusters (BGCs) encoding non-ribosomal peptide synthetases (NRPS), polyketide synthases (PKS), NRPS-PKS hybrids, or other BGC classes. It was applied to entomopathogenic bacteria of the genera Xenorhabdus and Photorhabdus by exchanging the natural promoter of desired BGCs against the L-arabinose inducible PBAD promoter in ∆hfq mutants of the respective strains. The crude (culture) extracts of the cultivated easyPACId mutants are enriched with the single compound or compound class and can be tested directly against various target organisms without further purification of the produced natural products. Furthermore, isolation and identification of compounds from these mutants is simplified due to the reduced background in the ∆hfq strains. The approach avoids problems often encountered in heterologous expression hosts, chemical synthesis, or tedious extraction of desired compounds from wild-type crude extracts. This protocol describes easyPACId for Xenorhabdus and Photorhabdus, but it was also successfully adapted to Pseudomonas entomophila and might be suitable for other proteobacteria that carry hfq.
Keywords: easyPACIdBackground
Entomopathogenic bacteria of the genera Xenorhabdus and Photorhabdus live in symbiosis with soil dwelling nematodes of the genera Steinernema or Heterorhabditis, respectively (Goodrich-Blair and Clarke, 2007; Chaston et al., 2011). They are able to propagate without their nematode host and can be easily cultivated in the laboratory using standard media. Their genome harbors a rich diversity for natural products (NPs) with potential activities as antibiotics, antifungals, and cytotoxic or signaling functions. These NPs are often produced by non-ribosomal peptide synthetases (NRPS), polyketide synthases (PKS), and mixed NRPS-PKS hybrids, encoded by biosynthetic gene clusters (BGCs) (H. B. Bode, 2009; Shi and Bode, 2018; Shi et al., 2022).
The access to this promising source of bioactive compounds is possible via different approaches: either by optimizing the culture conditions for enhanced NP production, by heterologous expression or promoter activation of the BGC of interest, or chemical synthesis of the NP derived thereof (H. B. Bode et al., 2002; Biggins et al., 2011; Biggins et al., 2014; E. Bode et al., 2017). All these methods have their limitations: chemical synthesis might be impossible, difficult, or too time consuming, requiring profound knowledge of the NP structure as a starting point. Heterologous production of the desired NP is dependent on building blocks that might only be present in the natural host, on the size of the BGC to be cloned into the heterologous host, on its toxicity, or on special transport systems or modifying enzymes (Brachmann et al., 2012; Nollmann et al., 2015). Isolation of natural products from crude extracts from wildtype (WT) bacteria might be hindered by low production titers, regulatory elements that silence the expression of BGCs, and/or sophisticated culture conditions that have to be established. A promoter activation of desired gene clusters in WT strains might result in NP overproduction that facilitates the isolation of the compound for bioactivity testing and structure elucidation, but still requires NP isolation from crude extracts. The easyPACId (easy Promoter Activation and Compound Identification) approach circumvents all these limitations.
easyPACId describes a simple and fast enrichment and preparation of selected NPs for bioactivity screening just from the crude extract of a bacterial culture of an induced ∆hfq_pCEP-gene_xyz easyPACId mutant. It avoids laborious steps often encountered during NP isolation from WT crude extracts and is not hindered by limitations concerning heterologous expression or chemical synthesis. The targeted activation via the PBAD promoter (Guzman et al., 1995) in a clean ∆hfq background serves as an excellent tool to enable production of only desired NPs or NP classes derived from single BGCs (Tobias et al., 2017; Valverde, 2017; E. Bode et al., 2019). The RNA chaperone Hfq is widespread in proteobacteria. It has a pleiotropic effect on gene expression by mediating the interaction of small non-coding RNAs (sRNA) and mRNAs, and thereby can activate or suppress translation (Vogel and Luisi, 2011). For NP-producing bacteria like Xenorhabdus, Photorhabdus, or Pseudomonas, a ∆hfq mutant has lost the ability to produce all or most NPs or shows a severely reduced NP amount. Consequently, the chromatogram of crude extracts from a ∆hfq mutant analyzed via HPLC/MS shows no NP-derived signals anymore (clean background) compared to HPLC/MS chromatograms of the corresponding WT where all NPs are produced, and their corresponding signals show a typical pattern for the strain (Figure 1a).
Briefly, the experiment starts via detection of potential BGCs encoding NRPSs, PKS, hybrids thereof, or other BGC classes, via antiSMASH analysis (or based on other bioinformatic tools) (Medema et al., 2011) in the genome of selected NP producers, here exemplified for the genera Photorhabdus and Xenorhabdus. By deletion of the gene hfq, encoding the so-called RNA-chaperone Hfq, a mutant is generated, which is deficient in NP production (Tobias et al., 2017). By exchange of the natural promoter against the L-arabinose inducible promoter PBAD (Guzman et al., 1995), encoded on the conjugatable vector pCEP via homologous integration (E. Bode et al., 2015), the selective activation of the targeted BGC in a ∆hfq background is now possible (Figure 1). Cell-free culture supernatants or even whole cultures from respective ∆hfq_pCEP-gene_xyz can be freeze dried and dissolved in water for subsequent testing on various target organisms (E. Bode et al., 2015 and 2019; Gulsen et al., 2022). Often, the selected compounds are overproduced and can be easily isolated for further investigations like structure elucidation.
This simple and beneficial method of gaining potentially bioactive compounds can be used to activate natural product biosynthesis gene clusters, not only in Xenorhabdus and Photorhabdus bacteria. It has been applied already to Pseudomonas and is likely to work in any other proteobacteria that encode Hfq, like Serratia or Vibrio (Thoma and Schobert, 2009; Hmelo et al., 2015). Besides Hfq itself, other regulative components of the hfq-sRNA regulon like ArcZ (Neubacher et al., 2020) or even other global regulators might also be useful to generate NP non-producing phenotypes in the described or other Gram-negative bacteria.
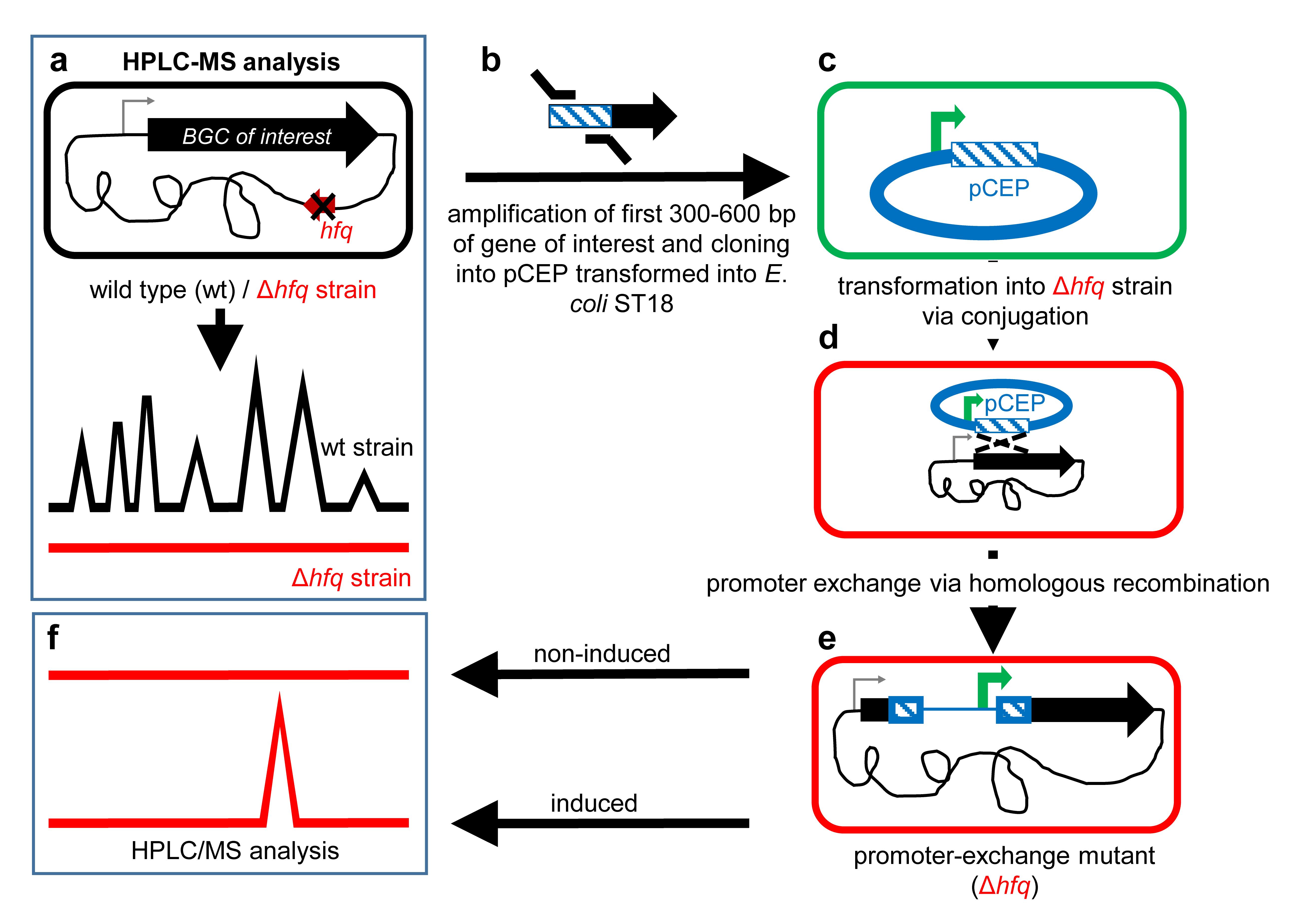
Figure 1. Schematic overview of the easyPACId workflow
Materials and reagents
∆hfq strain from Xenorhabdus spp. (E. Bode et al., 2019)
Sterile tips for micropipettes [Starlab, catalog numbers: S1110-3700-c (10–20 μL), S1111-1706-c (200 μL), S1111-6000-c (1,000 μL)]
Sterile reaction tubes [Sarstedt, catalog numbers: 72.706 (1.5 mL), 72.695.500 (2.0 mL)]
Black reaction tubes (Carl Roth, catalog number: AA80.1)
PCR tubes (Starlab, catalog number: 1402-3700)
50 mL tubes (Sarstedt, catalog number: 62.547.254)
Petri dishes [Sarstedt, catalog numbers: 82.1473.001 (92 mm × 16 mm), 82.1184.500 (150 mm × 20 mm)]
Electroporation cuvettes (Bio-Rad, catalog number: 1652089)
Single-use syringe Omnifix LuerLock Solo (Braun, catalog number: 4617207V)
Filtropur S 0.2 (Sarstedt, catalog number: 83.1826.001)
Steritop 45 mm neck size Millipore Express Plus 0.22 μm PES (Millipore, catalog number: S2GPT05RE)
Tryptone (BD, catalog number: 211699)
Yeast extract (BD, catalog number: 288620)
Sodium chloride (NaCl) (Carl Roth, catalog number: 3957.2)
Bacto-agar (BD, catalog number: 214030)
LE agarose (Biozym, catalog number: 840004)
L-Arabinose (Carl Roth, catalog number: 5118.5)
Sodium hydroxide (NaOH) (Carl Roth, catalog number: 6771.1)
Kanamycin monosulfate (km) (Sigma, catalog number: K4000)
5-Aminolevulinic acid hydrochloride (5-ALA) (Sigma, catalog number: 8339)
E. coli ST18 competent cells (Thoma and Schobert, 2009) (DSMZ, Braunschweig, Germany)
Conjugatable cloning vector pCEP-kmR with R6K ori, PBAD promoter, selection marker kanamycin (kmR) (E. Bode et al., 2015 and 2019)
Phusion High-Fidelity DNA Polymerase (New England Biolabs, catalog number: M0530L)
dNTP-Set (Carl Roth, catalog number: K039.2)
PstI/PstI HF (New England Biolabs, catalog numbers: R0140L/R3140L)
NdeI (New England Biolabs, catalog number: R0111L)
Monarch Genomic DNA Purification kit (New England Biolabs, catalog number: T3010L)
100 bp DNA ladder (New England Biolabs, catalog number: N3231L); 1 kb DNA ladder (New England Biolabs, catalog number: N3232L)
Midori Green Advance (Biozym, catalog number: 617004)
QIAprep Spin Miniprep kit (Qiagen, catalog number: 27106)
Wizard® SV Gel and PCR Clean-Up System (Promega, catalog number: A9285)
NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, catalog number: E2621L)
Methanol optima LC/MS grade (Fisher Scientific, catalog number: A456-212)
Ethanol (Carl Roth, catalog number: 9065.4)
Glycerol (Carl Roth, catalog number: 3783.2)
Acetic acid (Carl Roth, catalog number: 3738.5)
Trizma-Base (Sigma, catalog number: T6066)
Na2-EDTA (Carl Roth, catalog number: 8043.2)
Luria-Bertani medium (LB) (see Recipes)
5-Aminolevulinic acid hydrochloride (5-ALA) (50 mg/mL) (see Recipes)
L-Arabinose (20% w/v) (see Recipes)
Kanamycin (50 mg/mL) (see Recipes)
TAE-buffer, pH 8.0 (see Recipes)
0.5 M EDTA pH 8.0 (see Recipes)
Equipment
Super Duty Erlenmeyer flasks, 50 mL (Omnilab, catalog number: 5425046)
500 mL Erlenmeyer flasks (Omnilab, catalog number: 5010476)
Cellulose plug F 17.0–18.5 mm (VWR International, catalog number: 391-0161)
Caps for flasks with neck Ø38 mm (Carl Roth, catalog number: K396.1)
Sterile glass pipettes
Gel chambers for agarose gel electrophoresis
Inoculation loop, Platin-Iridium (neoLab, catalog number: 1-2112)
Drigalski spatula (Carl Roth, catalog number: T724.2)
Sterile toothpicks
Conical centrifuge tube 50 mL (Sarstedt, catalog number: 62.547.254)
ChemiDoc MP imaging systems with Lab Touch Software (Bio-Rad, catalog number: 12003154) and Blot/UV/Stain-Free tray (Bio-Rad, catalog number: 12003028)
Transilluminator (Intas Science Imaging Instruments)
Micropipettes (Gilson, Pipetman G P2G, catalog number: F144054M; Pipetman G P10G, catalog number: F144055M; Pipetman G P20G, catalog number: F144056M; Pipetman G P200G, catalog number: F144058M; Pipetman G P1000G, M, catalog number: F144059M)
Centrifuge Pico 17 microcentrifuge (Fisher Scientific, catalog number: 75002410)
Centrifuge Eppendorf 5810 R (V8.8), rotor S-4-104 with adapter (Eppendorf, catalog numbers: 5825734009 and 5825733002)
ProFlexTM PCR system, 96-well (Applied BiosystemsTM, catalog number: 4484075)
Eppendorf ThermoMixer C (Eppendorf, catalog number: 5382000015)
Infors HT-Multitron Standard shaker
Gene Pulser Xcell (Bio-Rad)
NanoDrop One WiFi (Life Technologies, catalog number: ND-ONE-W)
UV-vis-spectrophotometer Biochrom Libra S60 (Omnilab, catalog number: 80-7000-12)
Lyovapor L-300 (Büchi)
HPLC vials X1000 Microflaschen ND9 (Fisher Scientific, catalog number: 11707597)
Lid (for HPLC vials) X1000 (Fisher Scientific, catalog number: 11787567)
HPLC/MS Thermo Scientific UltiMate 3000 coupled to an amaZon speed ion trap (Bruker)
Vortex Genie 2 (Fisher Scientific, catalog number: 15547335)
Software
Geneious Prime® 2022.1.1 or earlier versions (https://www.geneious.com)
DataAnalysis (Bruker Daltonics GmbH)
antiSMASH (https://antismash.secondarymetabolites.org/#!/start) (Medema et al., 2011)
Procedure
Plasmid planning and construction
Choose the ∆hfq strain of interest and scan the genome for natural product BGCs using antiSMASH (Medema et al., 2011). This protocol will focus on Xenorhabdus_∆hfq as model organism for Xenorhabdus and Photorhabdus. For generating a ∆hfq strain in other proteobacteria, please refer to suitable methods for strain generation.
Streak the ∆hfq strain on a LB agar plate using a sterile inoculation loop in order to obtain single colonies. Incubate the inoculated plate at 30 °C for at least 72 h; look daily for growth and examine the strain for antibiotic resistance, pigmentation, swarming, or other phenotypical characteristics.
Note: For swarming strains, increase agar concentration of solid medium up to 30 g/L.
Store the grown plates at 18 °C for up to two weeks. For long-term storage, prepare glycerol stocks from fresh overnight cultures [720 μL of glycerol (50% v/v) and 1,280 μL of culture].
Inoculate 5 mL of LB medium in a 50 mL Erlenmeyer flask with a single colony from the plate and incubate the culture overnight at 30 °C rotating at 200 rpm. Purify chromosomal DNA from 1 mL (~2 × 109 cells) of bacterial culture with Monarch Genomic DNA Purification kit. Dissolve the DNA pellet in 100 μL of elution buffer or dH2O. Store dissolved DNA at 4 °C during the experimental period or freeze it at -20 °C for long-term storage.
Streak E. coli ST18_pCEP-kmR (Cluster Expression Plasmid) on LB agar containing 5-ALA (50 μg/mL) and kanamycin (50 μg/mL). Incubate overnight at 37 °C. Store the plates at 4 °C for up to two months. For long-term storage, follow step A3.
Note: The plasmid is also available as pCEP-cmR (E. Bode et al., 2015). This protocol will focus on pCEP-kmR.
Inoculate 10 mL of LB medium supplemented with 5-ALA (50 μg/mL) and kanamycin (50 μg/mL) with a single colony and cultivate the cells overnight at 37 °C shaking at 200 rpm.
Note: Always add 5-ALA (50 μg/mL) for cultivation of E. coli ST18 to the media, except in step C9.
Isolate pCEP plasmid DNA (Figure 2) (low copy vector) from 10 mL of culture (E. Bode et al., 2015), following the manual instruction of Qiagen Plasmid Mini Prep kit.
Note: For higher yield of the low copy vector pCEP, extract the vector from E. coli EC100 λpir cells, cultivate in SOC medium, or use Qiagen Midi Prep kit and extract plasmid DNA from 100 mL of overnight culture.
The cloning strategy is based on the HiFi DNA assembly method and carried out with the NEBuilder HiFi DNA Assembly Master Mix following the instruction manual.
Use Geneious Prime® and design a primer pair of 20–25 bp targeting the first 300–600 bp (homologous region) of the gene of interest (Figure 1b) with homologous arms (overlaps) (Table 1) to the pCEP vector (Figure 2c). For later validation of the promoter exchange in Xenorhabdus_∆hfq, design an individual verification primer Vxyz-rv binding approximately 20–100 bp downstream of the homologous region (Figure 3). Its melting temperature (Tm) should match the Tm of the verification primer VpCEP-fw that binds to the pCEP vector (Table 2, Figure 3).
Perform PCR on the genomic DNA, extracted in step A4, with Phusion polymerase. Amplify the first 300–600 bp of the target gene (Figure 1b). Run PCR on an agarose gel (1% agarose in TAE-buffer, stain with Midori Green Advance 0.5 µL/100 mL). Use the 100 bp DNA ladder for determination of the right fragment size and cut the appropriate DNA fragment out of the gel. Use Wizard® SV Gel and PCR Clean-Up System and extract the fragment from the gel slice.
Note: Other polymerases can be used for amplification, especially for GC-rich DNA. If no fragment is obtained via PCR, sometimes a 1:10 dilution of genomic DNA with deionized H2O is helpful. An optimization of the PCR reaction mixture by adding more DMSO or additional MgCl2 might be beneficial. Adjust the PCR protocol and cycle to the respective polymerase.
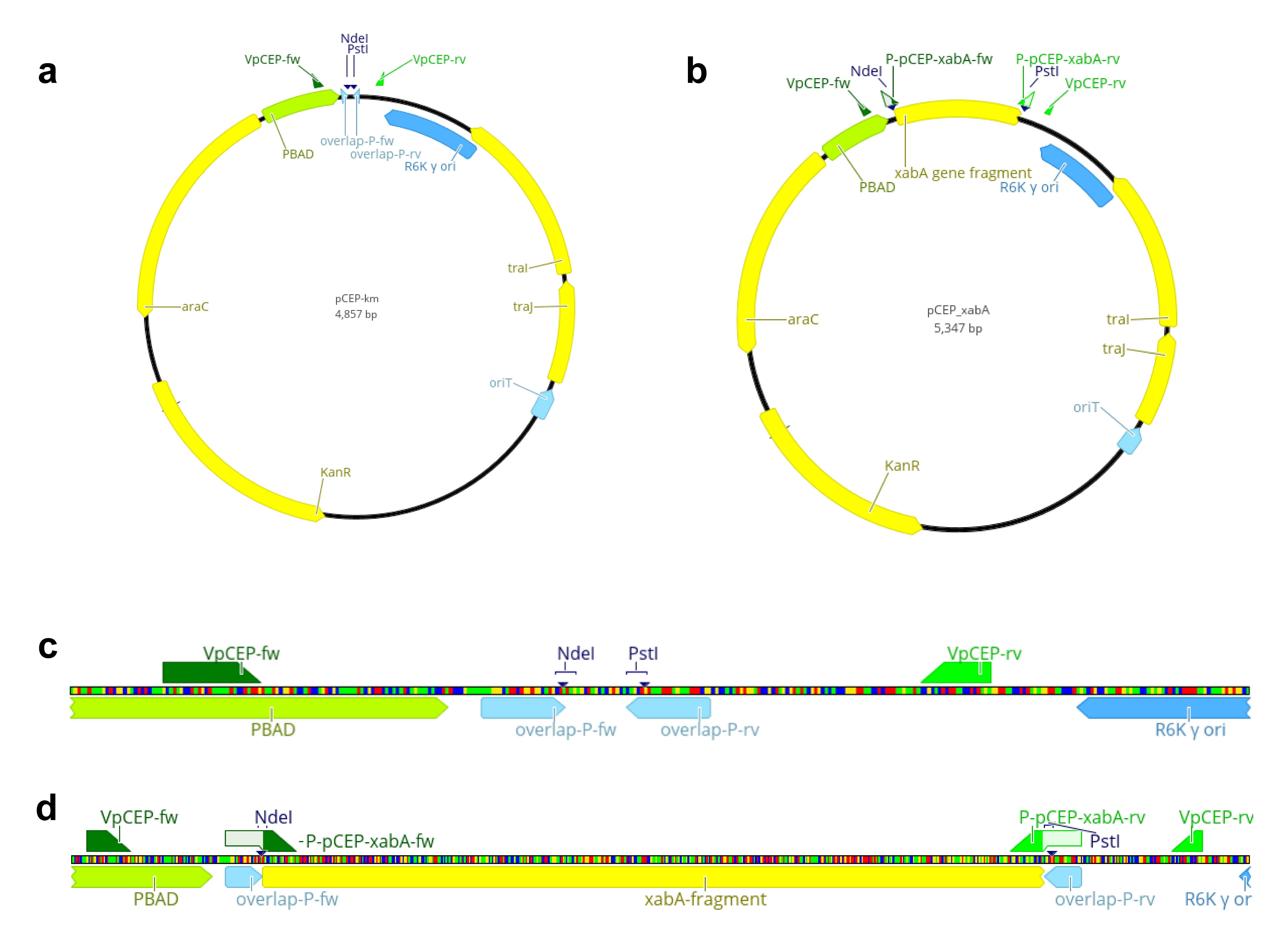
Figure 2. Schematic overview of the multiple cloning site of pCEP-km and pCEP_xabA. Vector map of pCEP-kmR (a) and pCEP_xabA (b), highlighting the empty multiple cloning site (MCS) and the restriction sites for the selected enzymes NdeI and PstI (c) and the inserted gene fragment xabA (d).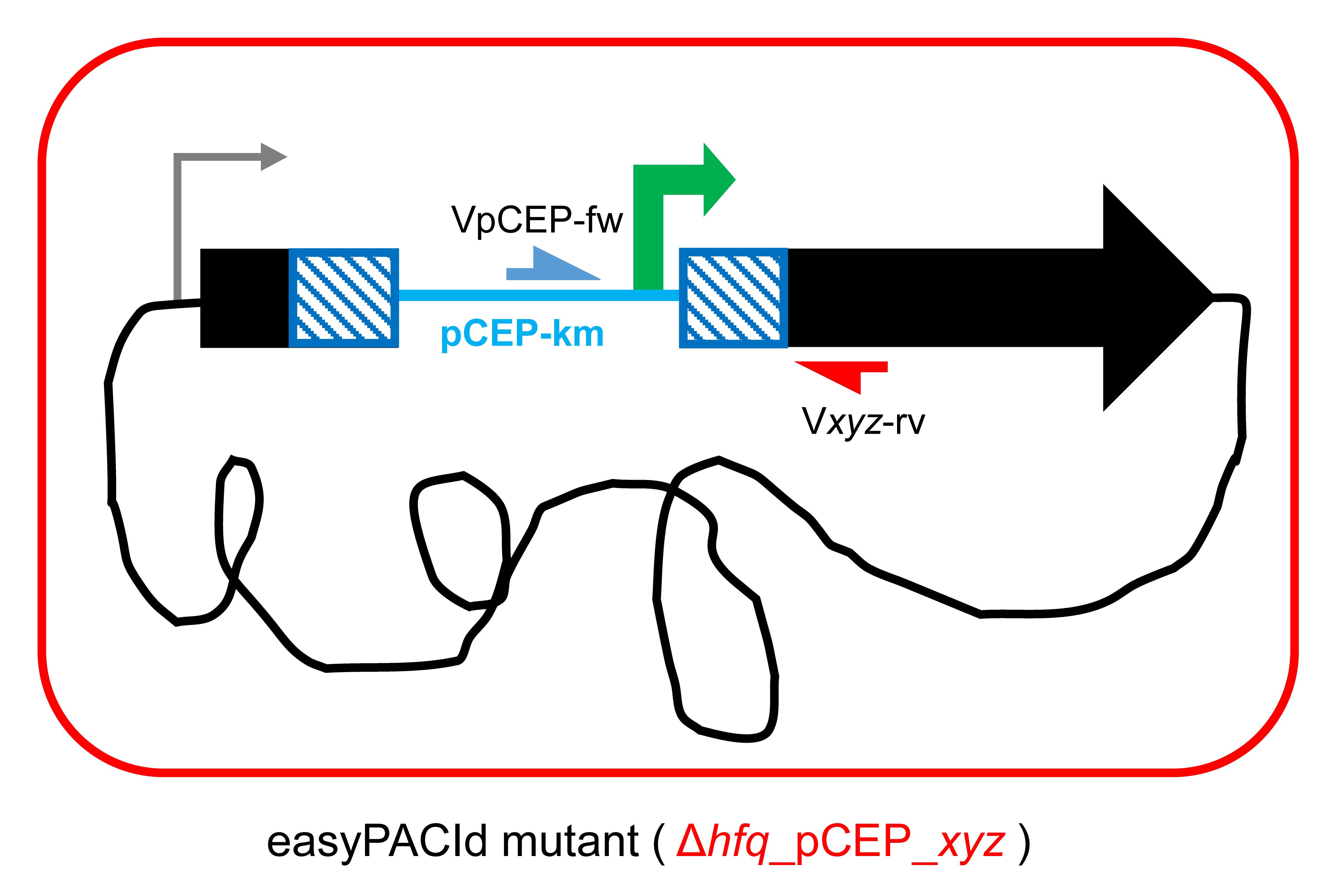
Figure 3. Schematic picture of “easyPACId mutant” Xenorhabdus_∆hfq_pCEP_xyz. The general verification primer VpCEP-fw (depicted in blue) binds to the integrated pCEP backbone, and the individual verification primer Vxyz-rv (depicted in red) binds in the genome, downstream of the homologous region.Table 1. Sequences for oligonucleotide-overlaps to the pCEP plasmid required for cloning/amplification of the gene of interest
Oligo Sequence 5′-3′ overlap-P-fw TTTGGGCTAACAGGAGGCTAGCAT_binding primer overlap-P-rv CCGTTTAAACATTTAAATCTGCAG_binding primer PCR protocol
1 μL
10 μL
1.5 μL
0.4 μL
0.5 μL
0.5 μL
1 μL
35.1 μL
50 μLgDNA
5× HF-buffer
DMSO
dNTPs (25 mM)
Primer forward (100 pmol/μL)
Primer reverse (100 pmol/μL)
Phusion-polymerase
H2O
volumePCR cycle
98 °C 2 min 98 °C 10 s 65 °C 30 s 35× 72 °C 30 s 72 °C 10 min 20 °C ∞ Perform a restriction of pCEP with enzymes NdeI and PstI (Figure 2). The restriction sites are located in the multiple cloning site of pCEP and enable the ligation of the prepared gene fragment via its homologous arms in front of the PBAD promoter, which is located upstream of the NdeI restriction site (Figure 2).
Run the restricted plasmid on an agarose gel (1% agarose in TAE buffer). Use 1 kb DNA ladder and determine the linear 4.84 kb plasmid backbone. Cut the fragment out of the gel and extract it from the gel slice with Wizard® SV Gel and PCR Clean-Up System.
Determine the DNA concentration of the plasmid backbone and insert fragments using a NanoDrop. Ligate the fragments using NEBuilder HiFi DNA Assembly Master Mix for 1 h at 50 °C, following the manual’s instructions. Additionally, set up a ligation mix that contains the restricted pCEP vector only and replace the volume of insert with dH2O as a control for restriction efficiency. After ligation, construct samples can be stored at -20 °C.
Note: Perform ligation in a PCR tube in a PCR thermocycler. If no clones are obtained, variate the vector/insert ratio.
Transformation and validation of plasmid constructs
Thaw the competent E. coli ST18 cells on ice. Transform 1–10 μL of the ligated construct (depending on the transformation method) into the donor cells via heat shock (a) or electroporation (b) (Figure 1b).
Thaw 50 μL of chemically competent cells on ice for 10 min. Add 5–10 μL of pCEP_xyz ligation mixture and incubate on ice for 30 min. Heat shock the cells for 45 s at 42 °C and subsequently incubate on ice for 2 min. Add 200 μL of LB with 5-ALA (50 μg/mL) medium. Let cells regenerate at 37 °C and 800 rpm in a ThermoMixer for 1 h.
Add 1–4 μL of pCEP_xyz ligation mixture to 50 μL of electro competent cells and mix well by pipetting up and down. Pipette the mixture onto the bottom of a pre-chilled electroporation cuvette (1 mm). Use Gene Pulser Xcell and pulse at 1.8 kV, 200 Ω, and 25 μF. Immediately add 950 μL of pre-warmed LB medium + 5-ALA (50 μg/mL), transfer the cells into a 1.5 mL reaction tube, and let cells recover at 37 °C shaking at 800 rpm in a ThermoMixer for 1 h.
Plate the transformed cells (Figure 1c) on LB agar containing 5-ALA and kanamycin. Incubate the agar plates overnight at 37 °C. Usually, colonies will appear the next day.
For cells transformed via heat shock: plate 100–200 μL of the transformed E. coli pCEP_xyz. Spin down the remaining culture at 17,000× g for 15 s, decant the supernatant, dissolve cells in the remaining liquid, and plate them on a second LB agar plate.
For electroporated cells: plate 100 μL on a first plate, then spin down the remaining culture at 17,000× g for 15 s, decant the supernatant, dissolve cells in the remaining liquid, and plate them on a second LB agar plate.
Screen for the accurate E. coli_pCEP-xyz donor by checking at least 10 single colonies by colony PCR: pick colonies with a sterile toothpick or sterile tip and suspend the cells in the colony PCR reaction mix. Set up a master plate with the corresponding selective antibiotic and 5-ALA by streaking each respective clone on the plate to save it for later applications.
Use verification primers VpCEP-fw and VpCEP-rv (Table 2) (Figure 2a and 2b) for amplification of the ligated fragment. A 234 bp fragment is the result for an empty vector.
Note: Alternatively, lyse colony material in 50 μL of dH2O and take 1 μL from the lysate for PCR. If the colony PCR gives no result, check your plasmid via restriction. Therefore, set up a small overnight culture from the respective mutant (5 mL in 50 mL flasks) in LB medium + antibiotic. Extract the plasmid from 5 mL of overnight culture and digest it with NdeI and PstI to obtain the ligated fragment size or use a single restriction enzyme that cleaves your plasmid twice.
Table 2. Oligonucleotide sequences for verification of the correct E. coli_pCEP_xyz clone binding upstream and downstream of the ligated fragment on the pCEP backbone (Figure 2)
Oligo Sequence 5′-3′ Tm VpCEP-fw GCTATGCCATAGCATTTTTATCCATAAG 64 °C VpCEP-rv ACATGTGGAATTGTGAGCGG 64 °C Colony PCR
1 μL
4 μL
0.6 μL
0.16 μL
0.2 μL
0.2 μL
0.5 μL
13.34 μL
20 μLCell material or 1 μL DNA lysate
5× HF-buffer
DMSO
dNTPs (25 mM)
Primer VpCEPforward (100 pmol/μL)
Verification primer (100 pmol/μL)
Phusion polymerase
dH2O
volume
PCR cycle98°C 3 min 98 °C 10 s 65 °C 30 s 35× 72 °C 30 s 72 °C 10 min 20 °C ∞
Mating procedure
Inoculate 5 mL of LB in a 50 mL flask with a single acceptor strain colony Xenorhabdus_∆hfq. Cultivate at 30 °C and 200 rpm overnight.
Note: For experiments with Xenorhabdus and Photorhabdus, the colonies should not be older than 10 days.
In parallel, inoculate the desired E. coli_pCEP-xyz donor in 5 mL of LB medium supplemented with 5-ALA and kanamycin in a 50 mL flask. Cultivate at 37 °C and 200 rpm overnight.
On the next day, inoculate the acceptor strain in 5 mL of fresh LB medium in two different ratios from the acceptor overnight culture—1:25 and 1:50—and incubate until OD600 = 0.6–1.0 at 30 °C while shaking at 200 rpm. Measure all OD600 with UV-vis-spectrophotometer Biochrom Libra S60.
In parallel, inoculate the donor E. coli_pCEP-xyz strain in 5 mL of LB medium + 5-ALA in two different ratios from the donor E. coli_pCEP-xyz overnight culture—1:50 and 1:100—and incubate until OD600 = 0.6–1.0 at 37 °C while shaking at 200 rpm. The OD is reached after 3–5 h.
Note: Prepare glycerol stocks from the overnight culture of the E. coli mutant for long-term storage. If the donor cells reach the appropriate OD earlier than the acceptor, store them on ice until the acceptor is ready.
When the OD600 is appropriate, pipette 1 mL from the acceptor strain into a reaction tube. Spin down the cells at 17,000× g for 1 min. Decant the supernatant.
Spin down 1 mL of the E. coli_pCEP_xyz donor at 17,000× g for 1 min.
Wash the E. coli_pCEP_xyz donor cells twice with 1 mL of LB medium to eliminate 5-ALA. Discard the supernatant.
Dissolve the pelleted cells in 400 μL of LB medium.
Combine donor and acceptor cells in a drop on an LB agar plate (do NOT add 5-ALA or antibiotics in this step) in the recommended acceptor:donor ratios: 1:3 (25 μL:75 μL) and 3:1 (75 μL:25 μL) (Figure 4a). Mix the cells in the drop by gently agitating the plate; do not spread the cells over the whole plate!
Incubate the plates for 3 h at 37 °C and subsequently switch to 30 °C for 21 h. Bacterial conjugation happens via the rolling circle mechanism. pCEP-xyz integrates via its homologous region “-xyz” into the genome of the acceptor strain, replacing the natural promoter by the inducible PBAD promoter (Figure 1d, Figure 3), generating the promoter exchange mutant ∆hfq_pCEP_xyz (Figure 1e, Figure 3).
Note: Do not exceed the incubation time at 37 °C as Xenorhabdus and Photorhabdus will not survive. If time is a problem, just incubate at 30 °C for 24 h, which also works well in most cases.
Selection for promoter exchange mutants
Scrape the cell plaque (Figure 4b) from the agar carefully with a sterile inoculation loop and suspend the cells in 1 mL of LB medium by pipetting up and down.
Inoculate the selection plate for ∆hfq_pCEP_xyz promoter exchange mutants by spreading 300 μL of the cell suspension on a large Petri dish containing LB agar + kanamycin (without 5-ALA!) by using a sterile Drigalski spatula or a glass pipette. Incubate the plates at 30 °C for at least 72 h and check for colonies daily (Figure 4c).
Note: Depending on the target gene, colonies may appear early, after 24 h of incubation, or late, after 72 h, if bacterial growth is severely affected by the silencing of the gene by promoter exchange.
Streak 1–10 single ∆hfq_pCEP_xyz colonies on an LB agar plate + selective antibiotic (master plate) (Figure 4d).
Perform colony PCR from selected cells: pick colony material from the plate and suspend the cells directly in the PCR samples, or dissolve material from the colony in 50 μL of dH2O or 25 μL of NaOH (0.02 M) in a PCR tube, and lyse cells at 99 °C for 5 min in the thermocycler to obtain genomic DNA. Take 1 μL for PCR and use VpCEP-fw (Table 2) and the corresponding individual verification primer Vxyz-rv (Figure 5) binding downstream of the homologous region in the genome, designed in step A9.
Note: If no fragment is obtained by colony PCR, extract genomic DNA described in step A for PCR.
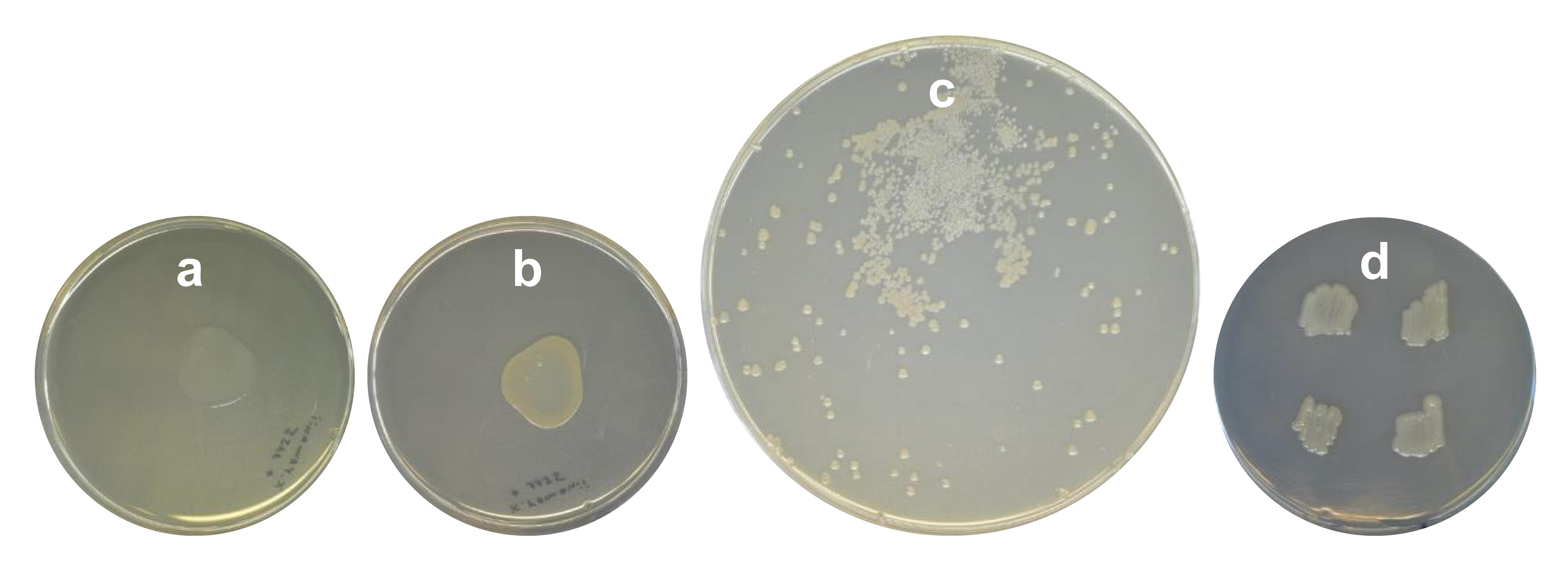
Figure 4. Different steps of conjugation. Fresh (a) and 24 h incubated (b) mating plaque of Xenorhabdus_∆hfq + E. coli ST18-pCEP_xyz, which will be scraped off and dissolved in LB (according to the protocol step D1). (c) Promoter exchange mutants Xenorhabdus_∆hfq_pCEP_xyz after plating on LB-km agar and 48 h of incubation at 30 °C. (d) Master plate on LB-km agar of selected mutants from plate depicted in (c).
Production cultures and HPLC/MS analysis
Inoculate 5 mL of LB + selective antibiotic in a 50 mL flask with a single ∆hfq_pCEP_xyz colony. Prepare respective cultures from the ∆hfq acceptor strain and the corresponding WT strain as controls.
Next day: wash the cells twice with LB medium to eliminate antibiotics.
Inoculate 100 mL of LB medium in 500 mL flasks with washed cells from the pre-cultures 0.1% (v/v) and induce all cultures with L-arabinose (final concentration 0.2%). Cultivate for 48–72 h at 30 °C and 140 rpm on a rotary shaker.
Transfer the cultures in 50 mL plastic tubes with a threaded cover and spin down the cells at 3,220× g for 20 min.
Note: Extend centrifugation time if the cells do not form a solid pellet.
Transfer the cell-free supernatant into new plastic tubes.
For HPLC/MS analysis: Pipette 50 μL of the supernatant in a reaction tube and extract it with 500 μL of methanol by mixing vigorously for 20 s on a vortex. Centrifuge the samples at 17,000× g for 20 min.
Pipette a sample of 50–120 μL into a HPLC vial for HPLC/MS analysis (Figure 5). Use DataAnalysis to analyze the HPLC/MS data.
Freeze the remaining cell-free supernatant at -80 °C.
Subsequently freeze dry the supernatant in a lyophilisator (Lyovapor L-300).
The lyophilizate can be stored at -20 °C for further investigations.
Dissolve the lyophilizate in 10 mL of deionized water and use the 10-fold concentrate for further analysis.
Note: Adjust solvents to specific testing conditions. To consider compounds that are located in the cell membrane, freeze dry the whole culture.
Data analysis
Exemplary data
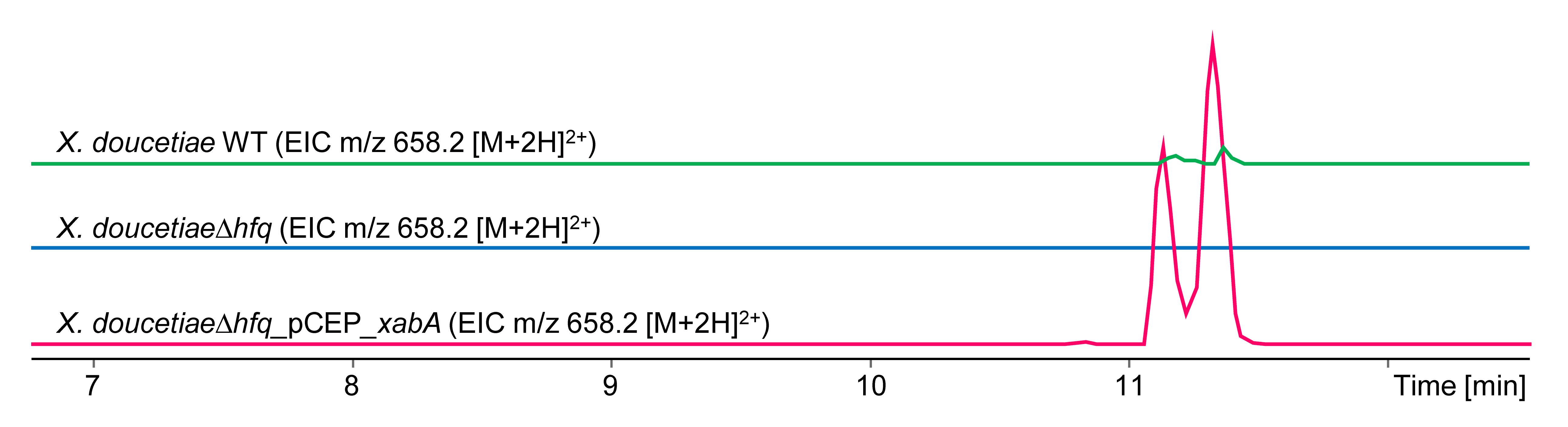
Figure 5. Extracted ion chromatograms (EICs) for xenoamicin (m/z 658.2 [M+2H]2+). Depicted are chromatograms from cultures in LB medium from X. doucetiae WT (green), X. doucetiae∆hfq (blue), and easyPACId mutant X. doucetiae∆hfq_pCEP_xabA (red) after induction with 0.2% L-arabinose. Cell-free supernatant was extracted with methanol according to the protocol step E6.
Recipes
Luria-Bertani medium (LB) (1 L)
Tryptone 10 g
Yeast extract 5 g
NaCl 5 g
dH2O 1 L
For solid medium, add
Bacto-agar 15 g
Sterilize by autoclaving for 20 min at 121 °C.
Note: Add antibiotics, 5-ALA, or L-arabinose after autoclaving.
5-Aminolevulinic acid hydrochloride (5-ALA) (50 mg/mL)
Prepare a stock solution with 50 mg/mL in dH2O, filter sterilize, and store at -20 °C in black reaction tubes to avoid photochemical degradation. Working concentration 50 μg/mL.
L-Arabinose (20% w/v)
Prepare a stock solution of 200 g/L in dH2O. Sterilize it by filtration via Steritop 45 mm neck size Millipore Express Plus.
Store at 4 °C or -20 °C for long-term storage.
Kanamycin (50 mg/mL)
Prepare a 50 mg/mL stock solution in dH2O, filter sterilize, and store at -20 °C.
TAE buffer, pH 8.0 (1 L)
Tris-Base 2.0 M 242 g
Acetic acid 2.0 M 57.1 mL
10% (v/v) EDTA 0.5 M pH 8.0 100 mL
0.5 M EDTA pH 8.0 (1 L)
Na2-EDTA·2H2O 182.1 g
Dissolve in ddH2O
Add 15 g of NaOH pellets and adjust pH with 5 M NaOH
Sterilize by autoclaving at 121 °C for 15 min
Acknowledgments
Part of this work was supported by an ERC Advanced Grant (835108) to H.B.B. The authors are grateful for support of the Max-Planck-Society. This protocol is derived from earlier versions described in the publications (E. Bode et al., 2015 and 2019).
Competing interests
The authors state that there are no competing interests.
References
- Biggins, J. B., Kang, H. S., Ternei, M. A., DeShazer, D. and Brady, S. F. (2014). The chemical arsenal of Burkholderia pseudomallei is essential for pathogenicity. J Am Chem Soc 136(26): 9484-9490.
- Biggins, J. B., Liu, X., Feng, Z. and Brady, S. F. (2011). Metabolites from the induced expression of cryptic single operons found in the genome of Burkholderia pseudomallei. J Am Chem Soc 133(6): 1638-1641.
- Bode, E., Brachmann, A. O., Kegler, C., Simsek, R., Dauth, C., Zhou, Q., Kaiser, M., Klemmt, P. and Bode, H. B. (2015). Simple “on-demand” production of bioactive natural products. Chembiochem 16(7): 1115-1119.
- Bode, E., He, Y., Vo, T. D., Schultz, R., Kaiser, M. and Bode, H. B. (2017). Biosynthesis and function of simple amides in Xenorhabdus doucetiae. Environ Microbiol 19(11): 4564-4575.
- Bode, E., Heinrich, A. K., Hirschmann, M., Abebew, D., Shi, Y. N., Vo, T. D., Wesche, F., Shi, Y. M., Grün, P., Simonyi, S., et al. (2019). Promoter Activation in Deltahfq Mutants as an Efficient Tool for Specialized Metabolite Production Enabling Direct Bioactivity Testing. Angew Chem Int Ed Engl 58(52): 18957-18963.
- Bode, H. B. (2009). Entomopathogenic bacteria as a source of secondary metabolites. Curr Opin Chem Biol 13(2): 224-230.
- Bode, H. B., Bethe, B., Höfs, R. and Zeeck, A. (2002). Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem 3(7): 619-627.
- Brachmann, A. O., Kirchner, F., Kegler, C., Kinski, S. C., Schmitt, I. and Bode, H. B. (2012). Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens. J Biotechnol 157(1): 96-99.
- Chaston, J. M., Suen, G., Tucker, S. L., Andersen, A. W., Bhasin, A., Bode, E., Bode, H. B., Brachmann, A. O., Cowles, C. E., Cowles, K. N., et al. (2011). The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS One 6(11): e27909.
- Goodrich-Blair, H. and Clarke, D. J. (2007). Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: two roads to the same destination. Mol Microbiol 64(2): 260-268.
- Gulsen, S. H., Tileklioglu, E., Bode, E., Cimen, H., Ertabaklar, H., Ulug, D., Ertug, S., Wenski, S. L., Touray, M., Hazir, C., et al. (2022). Antiprotozoal activity of different Xenorhabdus and Photorhabdus bacterial secondary metabolites and identification of bioactive compounds using the easyPACId approach. Sci Rep 12(1): 10779.
- Guzman, L. M., Belin, D., Carson, M. J. and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177(14): 4121-4130.
- Hmelo, L. R., Borlee, B. R., Almblad, H., Love, M. E., Randall, T. E., Tseng, B. S., Lin, C., Irie, Y., Storek, K. M., Yang, J. J., et al. (2015). Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10(11): 1820-1841.
- Medema, M. H., Blin, K., Cimermancic, P., de Jager, V., Zakrzewski, P., Fischbach, M. A., Weber, T., Takano, E. and Breitling, R. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39(Web Server issue): W339-346.
- Neubacher, N., Tobias, N. J., Huber, M., Cai, X., Glatter, T., Pidot, S. J., Stinear, T. P., Lutticke, A. L., Papenfort, K. and Bode, H. B. (2020). Symbiosis, virulence and natural-product biosynthesis in entomopathogenic bacteria are regulated by a small RNA. Nat Microbiol 5(12): 1481-1489.
- Nollmann, F. I., Dauth, C., Mulley, G., Kegler, C., Kaiser, M., Waterfield, N. R. and Bode, H. B. (2015). Insect-specific production of new GameXPeptides in photorhabdusluminescens TTO1, widespread natural products in entomopathogenic bacteria. Chembiochem 16(2): 205-208.
- Shi, Y. M. and Bode, H. B. (2018). Chemical language and warfare of bacterial natural products in bacteria-nematode-insect interactions. Nat Prod Rep 35(4): 309-335.
- Shi, Y. M., Hirschmann, M., Shi, Y. N., Ahmed, S., Abebew, D., Tobias, N. J., Grün, P., Crames, J. J., Pöschel, L., Kuttenlochner, W., et al. (2022). Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria. Nat Chem 14(6): 701-712.
- Thoma, S. and Schobert, M. (2009). An improved Escherichia coli donor strain for diparental mating. FEMS Microbiol Lett 294(2): 127-132.
- Tobias, N. J., Heinrich, A. K., Eresmann, H., Wright, P. R., Neubacher, N., Backofen, R. and Bode, H. B. (2017). Photorhabdus-nematode symbiosis is dependent on hfq-mediated regulation of secondary metabolites. Environ Microbiol 19(1): 119-129.
- Valverde, C. (2017). Who's the boss here? The post-transcriptional global regulator Hfq takes over control of secondary metabolite production in the nematode symbiont Photorhabdus luminiscens. Environ Microbiol 19(1): 21-24.
- Vogel, J. and Luisi, B. F. (2011). Hfq and its constellation of RNA. Nat Rev Microbiol 9(8): 578-589.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Bode, E., Assmann, D., Happel, P., Meyer, E., Münch, K., Rössel, N. and Bode, H. B. (2023). easyPACId, a Simple Method for Induced Production, Isolation, Identification, and Testing of Natural Products from Proteobacteria. Bio-protocol 13(13): e4709. DOI: 10.21769/BioProtoc.4709.
Category
Microbiology > Microbial physiology
Biological Engineering > Synthetic biology
Molecular Biology > DNA
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








