- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Cycloheximide (CHX) Chase Assay to Examine Protein Half-life
Published: Vol 13, Iss 11, Jun 5, 2023 DOI: 10.21769/BioProtoc.4690 Views: 16660
Reviewed by: Ralph Thomas BoettcherDeepali BhandariAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In-Gel Activity Assay of Mammalian Mitochondrial and Cytosolic Aconitases, Surrogate Markers of Compartment-Specific Oxidative Stress and Iron Status
Wing-Hang Tong and Tracey A. Rouault
Dec 5, 2024 2208 Views

Ub-POD: A Ubiquitin-Specific Proximity-Dependent Labeling Technique to Identify E3 Ubiquitin Ligase Substrates in Human Cells
Urbi Mukhopadhyay [...] Sagar Bhogaraju
Jun 20, 2025 2449 Views

Reliable and Sensitive Detection of Carbonylated Proteins by Oxime Blot
Filip Luka Mikulić [...] Mladen Merćep
Aug 5, 2025 1242 Views
Abstract
Cycloheximide (CHX) is a small molecule derived from Streptomyces griseus that acts as fungicide. As a ribosome inhibitor, CHX can restrict the translation elongation of eukaryotic protein synthesis. Once protein synthesis is inhibited by CHX, the level of intracellular proteins decreases by degradation through the proteasome or lysosome system. Thus, the CHX chase assay is widely recognized and used to observe intracellular protein degradation and to determine the half-life of a given protein in eukaryotes. Here, we present a complete experimental procedure of the CHX chase assay.
Graphical overview

Background
Regulation of protein abundance is crucial for the maintenance of normal cell function; all proteins in the cell are constantly being degraded and replaced. Specifically, rapid protein degradation plays an important role in regulating protein abundance in a variety of cellular processes, including cell cycle, signal transduction, and differentiation (Goldberg, 2003). Studies on protein degradation rate and half-life are important for understanding protein function, developing drugs to regulate the degradation of disease-specific proteins, and researching small molecule compounds to regulate protein levels (Paiva and Crews, 2019). There are two main protein degradation pathways in eukaryotic cells—the ubiquitin-proteasome pathway and the autophagy-lysosome pathway. The former is the main pathway of protein degradation in cells and is involved in the degradation of various proteins (Glickman and Ciechanover, 2002). It degrades proteins in cells in a specific, highly complex mechanism, and can be reversed by deubiquitinating enzymes (Sun et al., 2020). Autophagy-lysosome pathway is a self-degradation process, which degrades proteins through lysosomal mediation, by coating them and forming an autophagic lysosome, to fulfill the metabolic needs of the cells. Both pathways play vital roles in maintaining the normal protein metabolism of cells, and abnormal protein degradation may lead to many diseases, especially neurodegenerative disorders and cancer (Huang et al., 2018; Park et al., 2020).
The half-life of proteins varies widely, from a few minutes to days, and different rates of protein degradation are important for protein function. The classical method for protein degradation detection is the pulse-chase assay (Fritzsche and Springer, 2014); this assay is based on the addition of radionuclide-labeled amino acids, such as 35S-labeled methionine, to cell culture medium. The change process of a specific protein in the cells can be detected by immunoblotting or other methods. However, this method uses isotopes, which have some negative effects on the health and safety of researchers, and the operation process is complicated. Another common approach to study protein degradation is the protein translation blocker cycloheximide (CHX), which we will describe in detail later. In recent years, mass spectrometry technology has developed to detect the degradation of the primary structure of proteins. Therefore, protein degradation detection by mass spectrometry can be used to study protein structure, function, and quality control (Spradlin et al., 2021). In addition, it has been reported that CuAAC (copper-catalyzed azide–alkyne cycloadditions) can be used to label newly synthesized proteins in cells and to study protein degradation (Lu et al., 2013). Besides, protein degradation can be studied through the addition of pharmacological inhibitors of the degradation pathway to cells (Hanzl and Winter, 2020). For example, proteasome inhibitors such as MG-132, epoxomicin, and bortezomib block protein degradation via the ubiquitin-proteasome pathway. Inhibitors such as chloroquine and bafilomycin A1 work by neutralizing the acidic pH in the lysosome, which is necessary for the function of lysosomal protease.
Here, we describe a method that inhibits protein synthesis by adding CHX to assess the degradation of the target protein over time. CHX is a global translational inhibitor that can block the eukaryotic ribosome translocation process (Schneider-Poetsch et al., 2010). CHX is a product of Streptomyces griseus that can be used to determine the half-life of proteins or enzymes in molecular biology. It is noteworthy that CHX can inhibit eukaryotic protein synthesis but not prokaryotic protein synthesis. The advantages of CHX are that it is easy to operate, does not require considering the effect of ongoing protein synthesis, and allows for high-throughput screening. On the other hand, its main disadvantages are i) blocking all protein translation without specificity, and ii) having high cytotoxicity, able to cause large-scale cell death after prolonged treatment, so it is not suitable for proteins with a slow degradation rate. Therefore, CHX needs to be selected according to the appropriate conditions.
CHX chase assay visualizes protein degradation kinetics through CHX treatment over different times and western blotting analysis. Briefly, cells are treated with CHX at a suitable concentration, and then harvested at different timepoints. Next, western blot is employed to analyze the expression levels of a given protein. A short half-life protein will decrease in abundance over time, while a long half-life protein could exhibit little change in abundance. Thus, the CHX chase assay visualizes protein degradation efficiently without the use of radioactive isotopes, which provides an easy and reliable means to observe the change of intracellular protein levels and therefore could contribute to the study of therapeutic targets for a range of diseases.
Materials and reagents
Materials
Cell culture dish, 100 mm diameter (NEST, catalog number: 704001)
12-well plate (NEST, catalog number: 712011)
1,000 μL blue tip (Biosharp, catalog number: BS-1000-T)
200 μL yellow tip (Biosharp, catalog number: BS-200-T)
10 μL clear tip (Biosharp, catalog number: BS-10-T)
1.5 mL tube (AXYGEN, catalog number: AXYMCT150CS)
0.6 mL tube (AXYGEN, catalog number: AXYMCT060C)
Medical X-ray film (FUJIFILM, catalog number: SUPER RX-N)
Cell scraper (BIOLGIX, catalog number: 70-1180)
Cells
A549 cells (American Type Culture Collection)
Antibodies and plasmids
Anti-Tyk2 antibody (Cell Signaling Technology, catalog number: 14193; 1:1,000)
Anti-AHI1 antibody (Santa Cruz, catalog number: sc-515382; 1:500)
Anti-tubulin antibody (Proteintech, catalog number: 66031-1-Ig; 1:3,000)
Goat anti-rabbit IgG (H+L) HRP (Bioworld, catalog number: BS13278; 1:10,000)
Goat anti-mouse IgG (H+L) HRP (Bioworld, catalog number: BS12478; 1:10,000)
shCtrl plasmid (RNAi-Ready pSIREN-RetroQ-ZsGreen vector)
shAHI1 plasmid
shAHI1(#1)-forward: 5'-CACCGCGGAGACATTATCCGAGTGTTCGAAAACACTCGGATAATGTCTCCG-3'
shAHI1(#1)-reverse: 5'-AAAACGGAGACATTATCCGAGTGTTTTCGAACACTCGGATAATGTCTCCGC-3'
shAHI1(#2)-forward: 5'-CACCGCCATATTGGTCCGACAGTTTCGAAAAACTGTCGGACCAATATGGC-3'
shAHI1(#2)-reverse: 5'-AAAAGCCATATTGGTCCGACAGTTTTTCGAAACTGTCGGACCAATATGGC-3'
Reagents
Dulbecco’s modified Eagle medium (DMEM basic medium) (Gibco, catalog number: C11995500BT)
Fetal bovine serum (FBS) (PAN, catalog number: P30-3302)
Penicillin-streptomycin (Biosharp, catalog number: BL505A)
0.25% trypsin-EDTA (Biosharp, catalog number: BL512A)
Cycloheximide (CHX) (Sigma, catalog number: C7698-1G)
SuperSignal West Dura Extended kit (Thermo Scientific)
LongTrans (UCallM, catalog number: TF07)
Dimethyl sulfoxide for cell culture (DMSO) (panreac applichem, catalog number: 67-68-5)
Bradford (Sangon Biotech, catalog number: C900164-0200)
Non-fat milk (Biosharp, catalog number: BS102-500g)
BSA (Solarbio, catalog number: A8020)
Serum-free cell freezing medium (NCM Biotech, catalog number: C40100)
KCl (Sangon Biotech, catalog number: 7447-40-7)
NaCl (Sangon Biotech, catalog number: 7647-14-5)
EDTA (Solarbio, catalog number: 9002-07-7)
NP40 (Solarbio, catalog number: 9016-45-9)
Inhibitor cocktail (Sigma, catalog number: 58914000)
NaF (Sangon Biotech, catalog number: 7681-49-4)
PMSF (Solarbio, catalog number: 329-98-6)
Acr-Bis (30%) (Biosharp, catalog number: BL513B)
TEMED (Beyotime, catalog number: SY728)
APS (Sangon Biotech, catalog number: 7727-54-0)
SDS (Solarbio, catalog number: 151-21-3)
Glycine (Solarbio, catalog number: 56-40-6)
Tris base (Solarbio, catalog number: 77-86-1)
Tween 20 (Sangon Biotech, catalog number: 9005-64-5)
β-mercaptoethanol (Sigma, catalog number: 60-24-2)
Isopropyl alcohol (Sangon, catalog number: A507048-0500)
PageRuler prestained protein ladder (Thermo Fisher Scientific, catalog number: 26616)
Na3VO4 (Sangon Biotech, catalog number: 13721-39-6)
Na2HPO4 (Solarbio, catalog number: 7558-79-4)
KH2PO4 (Solarbio, catalog number: 7778-77-0)
Bromophenol blue (Solarbio, catalog number: 34725-61-6)
Glycerol (Solarbio, catalog number: G8190-500 ml)
HCl (Yonghua, catalog number: 210101204)
10× PBS Buffer (see Recipes)
Lysis buffer (1% NP40) (see Recipes)
8% separation gel (see Recipes)
5% stacking gel (see Recipes)
10× running buffer (see Recipes)
10× transfer buffer (see Recipes)
100 mM PMSF (see Recipes)
10% APS (see Recipes)
Stacking mix (see Recipes)
3× loading buffer (see Recipes)
SDS-sample buffer (see Recipes)
1× PBST buffer (see Recipes)
1% NP40 washing buffer (see Recipes)
1 M Tris-HCl (see Recipes)
Equipment
37 °C, 5% CO2 forced-air incubator (Thermo Scientific, catalog number: 3111)
Electric thermostatic water tank (Shanghai Jing Hong, catalog number: DK-8D)
Dry thermostat (AOSHENG, catalog number: MK-10)
SDS-PAGE gel casting apparatus (Tanon, catalog number: VE-180)
Protein electrophoresis and blotting instruments (Tanon, catalog number: EPS 300)
Centrifuge (Eppendorf, catalog number: 5425R)
Software
Microsoft PowerPoint
ImageJ
GraphPad Prism
Procedure
CHX can be used to determine changes in protein stability/half-live between different conditions, such as knockout vs. wild-type, overexpressed or in a knock-down situation. This protocol uses the example of Tyk2 stability in AHI1 knock-down cells.
Cell culture and transfection
Cells were cultured at 37 °C under 5% CO2 in DMEM supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin (complete medium). Cells were cultured with 0.25% trypsin-EDTA enzyme digesting technique and passaged in vitro.
Take A549 cells from -80 °C or liquid nitrogen and quickly place them in a 37 °C constant temperature water bath for approximately 2–3 min until the cells are completely thawed.
Place the cells into a centrifuge at 157× g for 3 min.
Remove the serum-free cell freezing medium, add 1 mL of complete medium to resuscitate cells, and transfer cells to a 100 mm culture dish.
Culture A549 cells in a 100 mm culture dish until 90% confluent.
Seed the cells at the ratio of 1:24 (5 × 105 cells) into six wells of a 12-well plate.
After 18–24 h, observe the cell density (approximately 80%–90%), and then perform the transfection of plasmids (such as shCtrl or shAHI1).
Remove the medium and add 1 mL of fresh complete medium before transfection.
Label 1.5 mL tubes as required and add 50 μL of DMEM basic medium to the tubes.
Prepare transfection solution A: add 1 μg of plasmid into DMEM medium and gently mix in the 1.5 mL tubes.
Prepare transfection solution B: add the LongTrans (DMEM:plasmid:longtrans = 50 μL:1 μg:2 μL) into DMEM medium and gently mix in the 1.5 mL tubes.
Gently mix transfection solutions A and B and leave at room temperature (RT) for 10–15 min.
Gently add the mixture to the corresponding medium and shake gently.
Place the plates in the incubator for approximately 36–48 h (for overexpressed plasmid) or 48–72 h (for knockdown plasmid).
Treat cells with CHX (50 μg/mL) for the appropriate timepoints (Section B).
CHX treatment
CAUTION: CHX is a highly toxic chemical and very toxic if swallowed; please avoid direct contact and release to the environment. In case of accident or if you feel unwell, seek medical advice immediately.
Remove the cell medium and replace with 1 mL of complete medium.
Distribute 500 μg of CHX into 0.6 mL tubes.
Add 10 μL of DMSO to the tube and mix well (the final concentration of CHX should be 50 mg/mL). CHX is better dissolved fresh to ensure maximal activity.
Slowly add 1 μL of CHX into the 1 mL cell culture medium and shake the plate gently to mix well. Similarly, add 1 μL of DMSO into the corresponding well as a control.
Harvest cells and extract cellular proteins (Section C).
Extracting cellular proteins
Prepare cell lysis buffer: add PMSF (1:100) to 1× NP-40 lysis buffer before use.
Place the cells on ice and aspirate the medium.
Gently add 1× PBS to wash and discard the PBS thoroughly. Repeat twice.
Add lysis buffer to the cells (100 μL per well).
Place the cells on ice for 30 min.
Scrap down the fully lysed cells with a cell scraper.
Transfer cell lysates to a 1.5 mL tube.
Place the 1.5 mL tubes into a 4 °C centrifuge at 12,000× g for 15 min.
Transfer the supernatant to a new 1.5 mL tube to get the cleared lysate (save at -80 °C); discard the cell debris sediment.
Use Bradford to determine protein concentration.
Denature 30–50 μg of proteins and add SDS-sample buffer to the samples.
Boil the samples in a metal bath at 96 °C for 10 min.
Centrifuge at 12,000× g for 1 min.
Use the samples for western blot or save them at -20 °C for future use.
Protein degradation analysis by western blot
Prepare the 8% SDS-PAGE gels (the gel concentration can be adjusted to 8%–12% depending on the molecular weight of the detected protein) with 8% separation gel and 5% stacking gel.
Dilute the 10× running buffer to 1× with double-distilled water (ddH2O).
Add 1× running buffer to the vertical electrophoresis chamber.
Gently pull out the comb of SDS-PAGE gel under running water and assemble it into vertical electrophoresis chamber.
Fill the chamber with running buffer.
Load the denatured protein samples into the lane of stacking gel and place 2 μL of PageRuler prestained protein ladder on both sides to indicate the molecular weight of the proteins.
Run for approximately 30 min using constant 80 V when the samples are in the stacking gel, and for approximately 90 min in the resolving gel at 110 V for a better separation of proteins.
Soak the PVDF membrane in absolute methanol for approximately 1 min to activate.
Prepare 1× transfer buffer (10× transfer buffer:absolute methanol:double-distilled water = 1:1:8).
Put the gel onto the activated PVDF membrane in 1× transfer buffer in the sequence of sponge, filter paper, gel, filter paper, and sponge.
Transfer the proteins to the gel using constant 360 mA for 90–120 min and immerse the electrophoresis tank in ice water.
Block the PVDF membrane with 5% non-fat milk with gentle shaking for 30 min at RT.
Dilute primary antibodies in 1% BSA at the indicated dilution.
Incubate the membrane with diluted primary antibodies with gentle shaking at RT for 2 h or 4 °C overnight.
Wash the membrane in 1× PBST buffer at RT for 10 min. Repeat three times.
Dilute the anti-mouse or anti-rabbit secondary antibodies in 5% non-fat milk at 1:10,000.
Incubate the membrane with the secondary antibodies with gentle shaking at RT for 1 h.
Wash the membrane in 1× PBST buffer at RT for 10 min. Repeat three times.
Visualize immunoreactive bands with a SuperSignal West Dura Extended kit using X-ray film in the dark room.
Edit gel images to the right size using Microsoft PowerPoint.
Use ImageJ to count pixels of the bands and calculate protein half-life.
Use GraphPad Prism to make the line chart.
Data analysis
Figure 1 displays a representative result showing the protein stability of Tyk2 in A549 cells transfected with shCtrl or shAHI1.
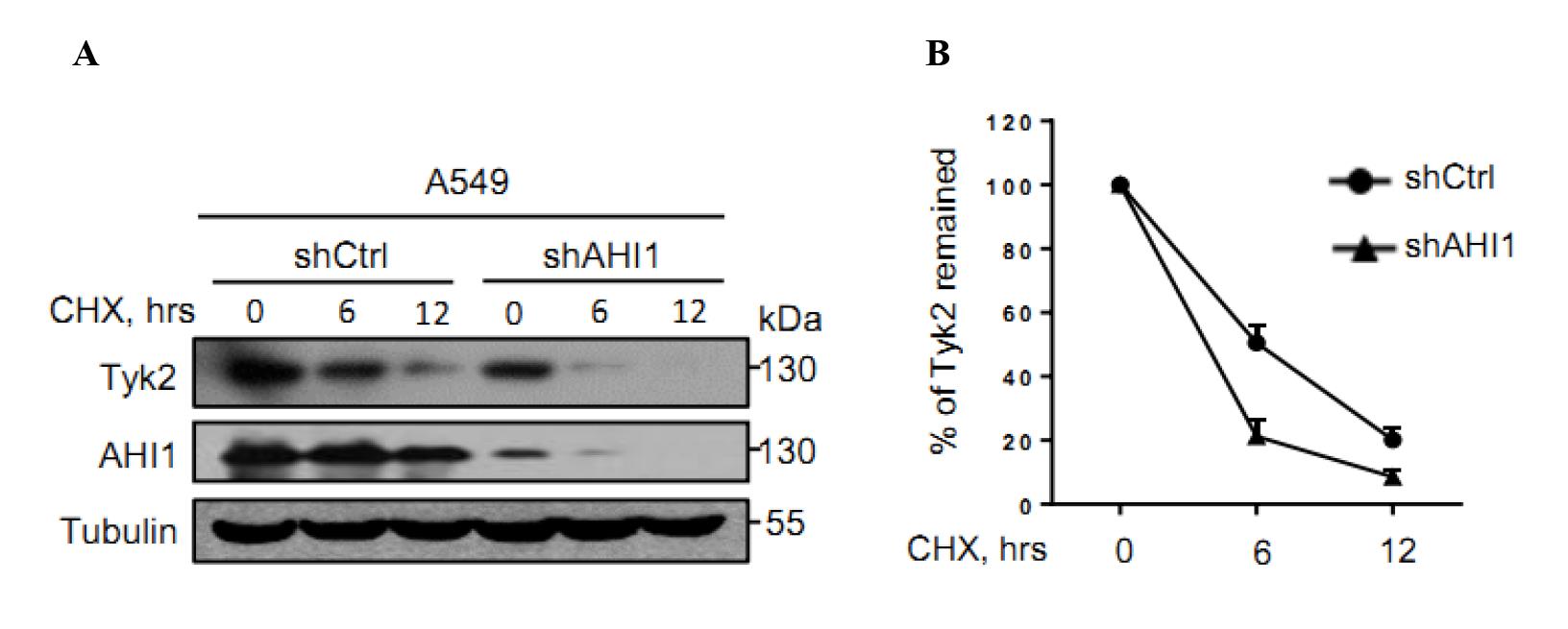
Figure 1. AHI1 knockdown accelerated Tyk2 protein degradation. (A) A549 cells were transfected with either control shRNAs (shCtrl) or shRNAs against AHI1 (shAHI1) plasmids. Seventy-two hours after transfection, cells were treated with either DMSO (for 0 h timepoint) or CHX (50 μg/mL, for 6 h and 12 h timepoints). Whole-cell extracts were analyzed by western blot using the antibodies against Tyk2, AHI1, or Tubulin. Tubulin serves as a loading control. (B) The striped images of Tyk2 and Tubulin were imported into Image J. The pixel count of all the strips was measured, and then the percentage of Tyk2 that remained was calculated in GraphPad Prism software. From Zhang et al. (2022).
Notes
The degradation rates of proteins can vary considerably from one another. Thus, it is suggested to determine the time course of protein degradation using the CHX chase assay in advance. Some endogenous proteins degrade very rapidly, and therefore the treatment time of CHX can be adjusted to 30–60 min. Sometimes it is necessary to set as few as five timepoints to detect the half-life of the protein. You can also find other experimental data about CHX chase assay in other papers of our research group (Feng et al., 2018; Yuan et al., 2020).
Recipes
10× PBS buffer
Reagent Amount (g) NaCl 80.00 KCl 2.00 Na2HPO4 14.20 KH2PO4 2.70 Dilute the 10× PBS buffer to 1× PBS with ddH2O.
Lysis buffer (1% NP40)
Reagent Volume (mL) 1% NP40 washing buffer (Recipe 13) 9.85 EDTA (0.5 M) 0.05 Na3VO4 (0.2 M) 0.05 NaF (0.5 M) 0.02 Inhibitor cocktail 0.02 8% separation gel
Reagent Volume (mL) ddH2O 3.80 Tris-HCl (pH 8.0) (Recipe 14) 2.00 Acr-Bis (30%) 2.13 10% APS (Recipe 8) 0.08 TEMED 8.00 × 10-3 5% stacking bel
Reagent Volume (mL) Stacking mix (see Recipe 9) 3.00 10% APS (Recipe 8) 0.02 TEMED 5.00 × 10-3 10× running buffer
Reagent Amount (g) Tris base 30.00 Glycine 144.00 SDS 40.00 Dilute the 10× running buffer to 1× running buffer with ddH2O
10× transfer buffer
Reagent Amount (g) Tris base 24.00 Glycine 142.00 Dilute the 10× transfer buffer to 1× transfer buffer with ddH2O
100 mM PMSF
Reagent Amount PMSF 0.696 g Isopropyl alcohol 40.00 mL 10% APS
Reagent Amount APS 1.00 g ddH2O 10.00 mL Stacking mix
Reagent Volume (mL) Acr-Bis (30%) 16.70 1 M Tris-HCl (pH 6.8) (Recipe 14) 12.50 20% SDS 0.50 ddH2O 70.30 3× loading buffer
Reagent Amount 1 M Tris-HCl (pH 6.8) (Recipe 14) 1.88 mL 20% SDS 3.00 mL Glycerol 3.00 mL Bromophenol blue 6.00 × 10-3 g ddH2O 2.20 mL SDS-sample buffer
Reagent Ratio (%) 3× loading buffer (Recipe 10) 85 β-mercaptoethanol 15 1× PBST buffer
Reagent Volume (mL) 1× PBS 500 Tween 20 5 1% NP40 washing buffer
Reagent Volume (mL) 5 M NaCl 1.50 1 M Tris-HCl (pH 7.4) 2.50 NP40 0.50 ddH2O 45.50 1 M Tris-HCl
Reagent Amount Tris 121.10 g HCl to pH = 6.8, 7.4, or 8.0 ddH2O to 1 L
Acknowledgments
This work was supported by grants from the Project of Natural Science Research in Colleges and Universities of Jiangsu Province (20KJB310008), the Foundation for Young Scholars of Jiangsu Province (BK20200864), and the National Key R&D Program of China (2018YFC1705500): 2018YFC1705505. This protocol was adapted from previous work published in Cell Research (Zhang et al., 2022).
Competing interests
The authors declare no conflict of interest.
Ethics
The study did not involve human subjects or animal work.
References
- Feng, Q., Miao, Y., Ge, J., Yuan, Y., Zuo, Y., Qian, L., Liu, J., Cheng, Q., Guo, T., Zhang, L., Yu, Z. and Zheng, H. (2018). ATXN3 Positively Regulates Type I IFN Antiviral Response by Deubiquitinating and Stabilizing HDAC3. J Immunol 201(2): 675-687.
- Fritzsche, S. and Springer, S. (2014). Pulse-chase analysis for studying protein synthesis and maturation. Curr Protoc Protein Sci 78: 30 33 31-30 33 23.
- Glickman, M. H. and Ciechanover, A. (2002). The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82(2): 373-428.
- Goldberg, A. L. (2003). Protein degradation and protection against misfolded or damaged proteins. Nature 426(6968): 895-899.
- Hanzl, A. and Winter, G. E. (2020). Targeted protein degradation: current and future challenges. Curr Opin Chem Biol 56: 35-41.
- Huang, T., Song, X., Yang, Y., Wan, X., Alvarez, A. A., Sastry, N., Feng, H., Hu, B. and Cheng, S. Y. (2018). Autophagy and Hallmarks of Cancer. Crit Rev Oncog 23(5-6): 247-267.
- Lu, B., Al-Ramahi, I., Valencia, A., Wang, Q., Berenshteyn, F., Yang, H., Gallego-Flores, T., Ichcho, S., Lacoste, A., Hild, M., et al. (2013). Identification of NUB1 as a suppressor of mutant Huntington toxicity via enhanced protein clearance. Nat Neurosci 16(5): 562-570.
- Paiva, S. L. and Crews, C. M. (2019). Targeted protein degradation: elements of PROTAC design. Curr Opin Chem Biol 50: 111-119.
- Park, H., Kang, J. H. and Lee, S. (2020). Autophagy in Neurodegenerative Diseases: A Hunter for Aggregates. Int J Mol Sci 21(9).
- Schneider-Poetsch, T., Ju, J., Eyler, D. E., Dang, Y., Bhat, S., Merrick, W. C., Green, R., Shen, B. and Liu, J. O. (2010). Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Cem Biol 6(3): 209-217.
- Spradlin, J. N., Zhang, E. and Nomura, D. K. (2021). Reimagining Druggability Using Chemoproteomic Platforms. Acc Chem Res 54(7): 1801-1813.
- Sun, T., Liu, Z. and Yang, Q. (2020). The role of ubiquitination and deubiquitination in cancer metabolism. Mol Cancer 19(1): 146.
- Yuan, Y., Miao, Y., Qian, L., Zhang, Y., Liu, C., Liu, J., Zuo, Y., Feng, Q., Guo, T., Zhang, L., et al. (2020). Targeting UBE4A Revives Viperin Protein in Epithelium to Enhance Host Antiviral Defense. Mol Cell 77(4): 734-747 e737.
- Zhang, H. G., Wang, B., Yang, Y., Liu, X., Wang, J., Xin, N., Li, S., Miao, Y., Wu, Q., Guo, T., et al. (2022). Depression compromises antiviral innate immunity via the AVP-AHI1-Tyk2 axis. Cell Res 32(10): 897-913.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Miao, Y., Du, Q., Zhang, H. G., Yuan, Y., Zuo, Y. and Zheng, H. (2023). Cycloheximide (CHX) Chase Assay to Examine Protein Half-life. Bio-protocol 13(11): e4690. DOI: 10.21769/BioProtoc.4690.
- Zhang, H. G., Wang, B., Yang, Y., Liu, X., Wang, J., Xin, N., Li, S., Miao, Y., Wu, Q., Guo, T., et al. (2022). Depression compromises antiviral innate immunity via the AVP-AHI1-Tyk2 axis. Cell Res 32(10): 897-913.
Category
Cell Biology > Cell metabolism > Other compound
Biochemistry > Protein > Degradation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









