- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
E15.5 Mouse Embryo Micro-CT Using a Bruker Skyscan 1172 Micro-CT
Published: Vol 13, Iss 9, May 5, 2023 DOI: 10.21769/BioProtoc.4662 Views: 1418
Reviewed by: Alberto RissoneEVANGELOS THEODOROUAbhilash Padavannil

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimizing Confocal Imaging Protocols for Muscle Fiber Typing in the Mouse Masseter Muscle
Catalina Matias [...] Jeffrey J. Brault
Apr 5, 2025 2916 Views

A Novel Optimized Silver Nitrate Staining Method for Visualizing and Quantifying the Osteocyte Lacuno-Canalicular System (LCS)
Jinlian Wu [...] Libo Wang
Apr 20, 2025 1532 Views

Examining the Roles of m6A Sites in mRNA Using the Luciferase Gene Fused With Mutated RRACH Motifs
Nobuko Katoku-Kikyo and Nobuaki Kikyo
Nov 5, 2025 1944 Views
Abstract
X-ray computed microtomography (µCT) is a powerful tool to reveal the 3D structure of tissues and organs. Compared with the traditional sectioning, staining, and microscopy image acquisition, it allows a better understanding of the morphology and a precise morphometric analysis. Here, we describe a method for 3D visualization and morphometric analysis by µCT scanning of the embryonic heart of iodine-stained E15.5 mouse embryos.
Keywords: ImagingBackground
Transgenic mouse models have been extensively used to study the roles of specific genes in heart development. Historically, to analyze morphological defects on heart formation in gain-of-function and loss-of-function models, the formalin-fixed paraffin-embedded embryos were sectioned and analyzed section-by-section. X-ray computed microtomography (µCT) is a powerful tool for visualizing the 3D morphology of the heart and performing morphometric analysis. µCT uses X-rays to create cross-sections of a physical object that can be used to recreate 3D models, reaching very high resolution (<1 µm). Here, we describe a method to apply this technique to biological samples. Our purpose was to study the cardiovascular system in the developing mouse embryo. This method overcomes the limits of traditional histology, such as the difficulty to obtain comparable sections, especially when working with very small and complex samples.
For µCT analysis of embryos, it is mandatory to increase the contrast of tissues. To this end, different methods and contrast agents are described in the literature, with small differences in protocols. The main agents available to this purpose and tested by the authors are iodine solution or phosphotungstic acid (PTA) (Wong et al., 2012 and 2013; Hsu et al., 2016; Handschuh and Glösmann, 2022). Osmium tetroxide is also used as a contrast agent, but because of its high toxicity it has not been tested by the authors. Both approaches of staining were previously used to study cardiac defects (Degenhardt et al., 2010; Lesciotto et al., 2020; Jamet et al., 2022). In our experience, the use of 2%–5% PTA solution requires methanol (3%–10%) to properly penetrate tissues. The concentration of PTA and methanol can be adjusted depending on the size of sample; for example, for small mouse embryos (E12.5), 3% of methanol should be sufficient; for E15.5 embryos, 5% methanol; and for large mouse embryos (E18.5), 10% of methanol should be necessary. We successfully used this method for the staining of E13.5 mouse embryos (Petrillo et al., 2018). Although the PTA method offers excellent contrast, it may cause artifacts due to sample shrinkage in embryos older than E13.5. The maximum size of the object suitable for the µCT is 50 mm; however, increasing sample size will reduce the resolution.
On the contrary, iodine-based staining offers good balance in terms of sample preservation and soft tissue contrast, especially for larger samples. In our experiment, in E15.5 embryos we used a 20% Lugol’s solution and 0.5% Tween-20. To preserve mineral structures, pH can be adjusted to 7.3.
Materials and Reagents
Mouse mating and embryo dissection
Petri dishes (100 mm) (BD Biosciences, catalog number: 351029 or similar)
35 mm cell culture dishes (BD Biosciences, catalog number: 352096 or similar)
Sterile disposable tubes (50 mL) (BD Biosciences, catalog number: 352070 or similar)
15 mL tubes
Mice
Pure ethanol (Sigma, catalog number: 493546)
Phosphate-buffered saline (PBS), without Ca2+ and Mg2+ (Life Technologies, Gibco®, catalog number: 10010)
4% paraformaldehyde (PFA) (Santa Cruz Biotechnology, catalog number: sc-281692)
70% ethanol (see Recipes)
Micro-CT sample preparation
Lugol’s solution (Sigma, catalog number: 32922)
Tween-20 (Sigma, catalog number: P1379)
Staining solution with contrast agent (see Recipes)
Equipment
Blunt forceps (FST Standard Pattern forceps or similar)
Fine forceps (Dumont #5 or similar) and scissors (FST, catalog number: 14060-09 or similar)
X-Ray computed microtomography Skyscan 1172 (Bruker)
This instrument is a desktop µCT equipped with a fully distortion-corrected 11 MP X-ray camera (a 12-bit cooled CCD camera coupled to scintillator). The X-ray source is 20–100 kV, 10 W, with a <5 µm spot size. 0.8 µm is the highest resolution (only for small samples) and 25 µm is the lower resolution. The maximum object size is 50 mm. Skyscan 1172 is equipped with three filter positions (None, 0.5 mm Al, or Al+Cu).
Software
DataViewer 1.5.4.6 (Bruker)
NRecon 1.4.4 (Bruker)
CTVox 3.3.0 (Bruker)
Procedure
Mice mating
Keep mice on a 12:12 h light/dark cycle with unrestricted access to food and water. Use animals aged 2–4 months for mating.
For mating, put a male and two females in a cage after 6 pm.
Before 9 am on the next day, check for the formation of vaginal plug: lift a female by her tail, slightly dilate the vaginal opening with a blunt forceps, and check for a white mass.
Consider the day of plug detection as day E0.5. House the mated females separately from the male. Repeat mating procedure with non-mated females the following days.
Embryo dissection
Embryo dissection is performed as described in Zeeb et al. (2012).
At day E15.5, euthanize the pregnant female.
Place the mouse on a supine position and spray 70% ethanol on the abdomen.
Make a V-formed incision on the skin and abdominal cavity and lift the tissue to expose the internal organs.
Identify the V-shaped uterus with the string of embryos inside, cut at the proximal (cervical) and distal (ovaries) ends, and remove the uterus to a tube with ice-cold PBS.
Dissect the embryos in a dish with PBS on ice. First, cut the uterus into single embryos-containing pieces. Place each embryo in a 35 mm dish with PBS and remove the uterine tissues and yolk sac. Keep yolk sac membranes for genotyping if required.
Embryo fixation
Place single embryos in 15 mL tubes with 5 mL of 4% PFA and incubate at 4 °C for 24 h.
After fixation, keep the embryos in PBS at 4 °C. If needed, the samples can be kept at 4 °C for several days.
Micro-CT sample preparation
Prepare fresh staining solution of 20% Lugol’s solution + 0.5% Tween-20 pH 7.3.
Keep samples in staining solution in constant gentle agitation.
Change solution daily for 10 days.
Micro-CT acquisition
Perform acquisition at 80 kV using a 0.5 mm Al filter at a resolution of 3 µm, 0.4° of rotation step, 360° scan, and 4× frame averaging.
Micro-CT image reconstruction
Obtain image reconstruction using NRecon software (Bruker), with proper misalignment compensation, no smoothing, 5 ring artifacts reduction, and 30% beam-hardening correction.
Data analysis
Open dataset of the single embryo in DataViewer software (Figure 1).
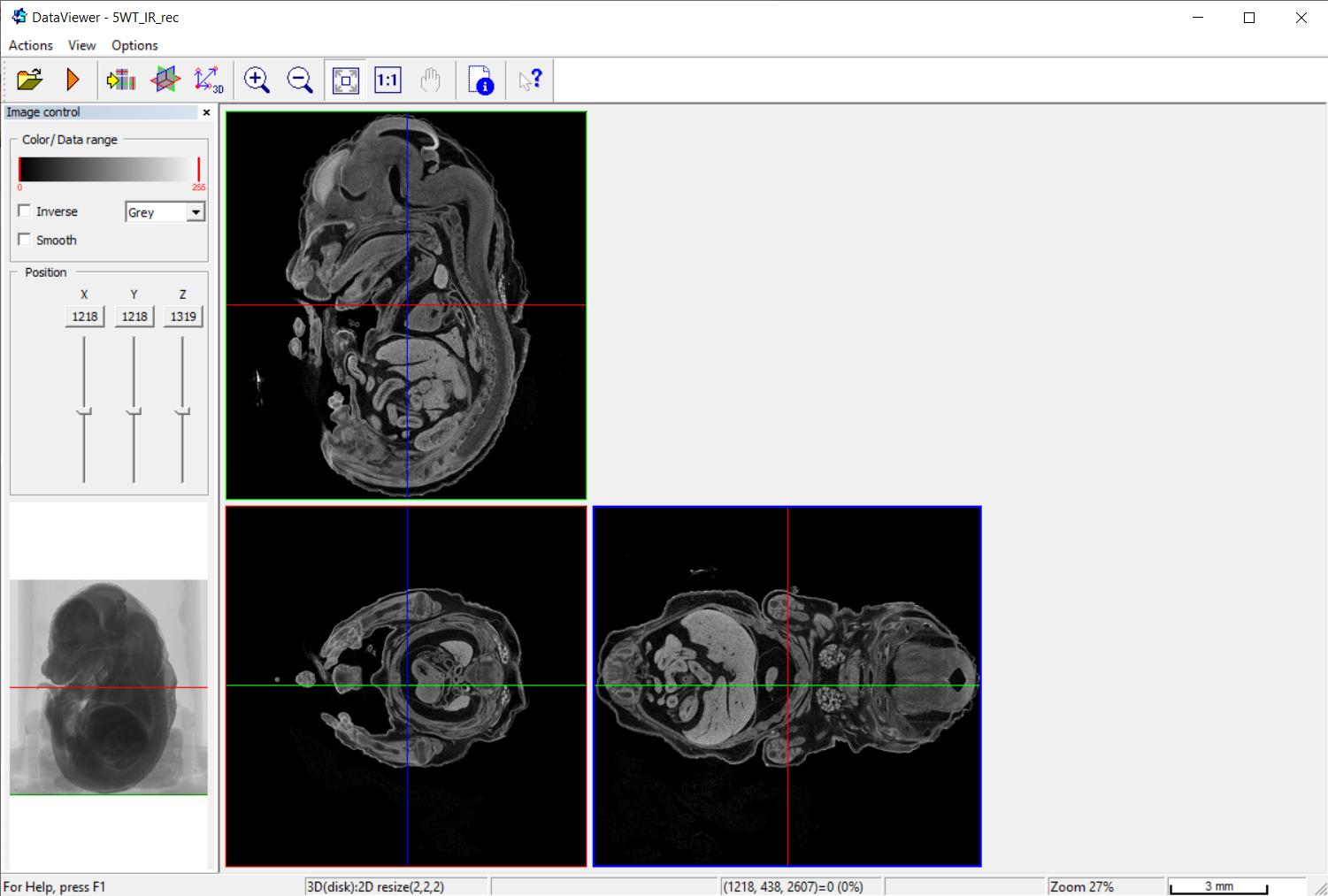
Figure 1. A screenshot of DataViewer software. An embryo is visualized in virtual sections in three planes.Select the plane by moving along the x- and y-axes. In our case, we were interested in performing the morphometric analysis of the embryo heart, in particular to measure the width of the ventricular walls and interventricular septum and the area of the pericardial cavity. The same plane, the best at representing the features we were interested in, was selected in all analyzed embryos.
Measure the features of interest by right-clicking to obtain the length in μm.
To perform a qualitative investigation, the dataset could be visualized in 3D by using CTVox software (Figure 2 and Video 1).
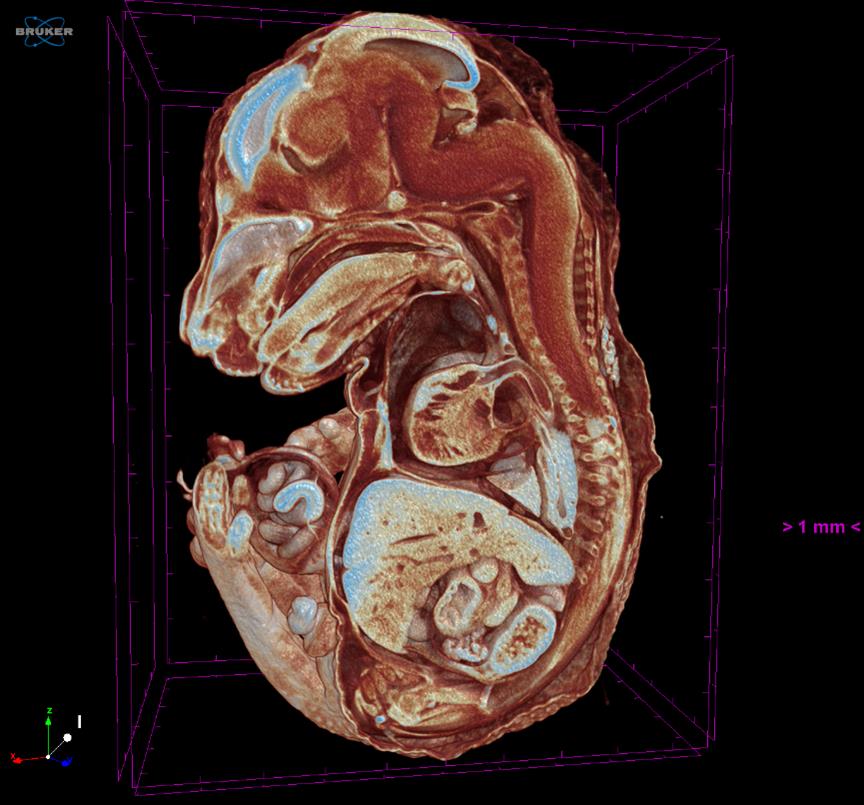
Figure 2. A screenshot of CTVox software. A E15.5 embryo is visualized in 3D slicing.Video 1. Embryo
Recipes
70% ethanol
Obtained by mixing 70 mL of pure ethanol with 30 mL of water.
Staining solution with contrast agent
Staining solution with contrast agent is obtained by making a 20% Lugol’s solution with 0.5% Tween-20 and adjusting pH to 7.3.
Acknowledgments
This protocol was adapted from the previously published study (Astanina et al., 2022). This work was supported by AIRC – Associazione Italiana Per la Ricerca sul Cancro (grants 22910), Regione Piemonte (grant A1907A, Deflect), Fondazione CRT, Ministero dell’Università e della Ricerca (PRIN 2017, grant 2017237P5X), FPRC 5xmille 2016 MIUR (Biofilm), and ERA-Net Transcan-2 (grant TRS-2018-00000689) to F.B. and Ministero dell’Università.
Competing interests
The authors declare no conflict of interests.
Ethics
All animal procedures were approved by the ethics committee of the University of Turin and by the Italian Ministry of Health (protocol approval no. 864/2015‐PR).
References
- Astanina, E., Doronzo, G., Corà, D., Neri, F., Oliviero, S., Genova, T., Mussano, F., Middonti, E., Vallariello, E., Cencioni, C., et al. (2022). The TFEB-TGIF1 axis regulates EMT in mouse epicardial cells. Nat Commun 13(1): 5191.
- Degenhardt, K., Wright, A. C., Horng, D., Padmanabhan, A. and Epstein, J. A. (2010). Rapid 3D phenotyping of cardiovascular development in mouse embryos by micro-CT with iodine staining. Circ Cardiovasc Imaging 3(3): 314-322.
- Handschuh, S. and Glösmann, M. (2022). Mouse embryo phenotyping using X-ray microCT. Front Cell Dev Biol 10: 949184.
- Hsu, C. W., Wong, L., Rasmussen, T. L., Kalaga, S., McElwee, M. L., Keith, L. C., Bohat, R., Seavitt, J. R., Beaudet, A. L. and Dickinson, M. E. (2016). Three-dimensional microCT imaging of mouse development from early post-implantation to early postnatal stages. Dev Biol 419(2): 229-236.
- Jamet, S., Ha, S., Ho, T. H., Houghtaling, S., Timms, A., Yu, K., Paquette, A., Maga, A. M., Greene, N. D. E. and Beier, D. R. (2022). The arginine methyltransferase Carm1 is necessary for heart development. G3 (Bethesda) 12(8).
- Lesciotto, K. M., Motch Perrine, S. M., Kawasaki, M., Stecko, T., Ryan, T. M., Kawasaki, K. and Richtsmeier, J. T. (2020). Phosphotungstic acid-enhanced microCT: Optimized protocols for embryonic and early postnatal mice. Dev Dyn 249(4): 573-585.
- Petrillo, S., Chiabrando, D., Genova, T., Fiorito, V., Ingoglia, G., Vinchi, F., Mussano, F., Carossa, S., Silengo, L., Altruda, F., et al. (2018). Heme accumulation in endothelial cells impairs angiogenesis by triggering paraptosis. Cell Death Differ 25(3): 573-588.
- Wong, M. D., Dorr, A. E., Walls, J. R., Lerch, J. P. and Henkelman, R. M. (2012). A novel 3D mouse embryo atlas based on micro-CT. Development 139(17): 3248-3256.
- Wong, M. D., Spring, S. and Henkelman, R. M. (2013). Structural stabilization of tissue for embryo phenotyping using micro-CT with iodine staining. PLoS One 8(12): e84321.
- Zeeb, M., Axnick, J., Planas-Paz, L., Hartmann, T., Strilic, B. and Lammert, E. (2012). Pharmacological manipulation of blood and lymphatic vascularization in ex vivo-cultured mouse embryos. Nat Protoc 7(11):1970-1982.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Astanina, E., Petrillo, S., Genova, T., Mussano, F. and Bussolino, F. (2023). E15.5 Mouse Embryo Micro-CT Using a Bruker Skyscan 1172 Micro-CT. Bio-protocol 13(9): e4662. DOI: 10.21769/BioProtoc.4662.
Category
Developmental Biology > Morphogenesis > Organogenesis
Cell Biology > Tissue analysis > Tissue imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









