- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protein Pull-down Assay Using HiBiT-tag-dependent Luciferase Activity Measurement
Published: Vol 13, Iss 6, Mar 20, 2023 DOI: 10.21769/BioProtoc.4640 Views: 2645
Reviewed by: Gal HaimovichKeisuke TabataAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Determination of Dissociation Constants for the Interaction of Myosin-5a with its Cargo Protein Using Microscale Thermophoresis (MST)
Rui Zhou [...] Xiang-Dong Li
Feb 5, 2025 1749 Views

Cell-Sonar, an Easy and Low-cost Method to Track a Target Protein by Expression Changes of Specific Protein Markers
Sabrina Brockmöller [...] Simone Rothmiller
Feb 5, 2025 1659 Views

SiMPull-POP: Quantification of Membrane Protein Assembly via Single Molecule Photobleaching
Ryan J. Schuck [...] Rajan Lamichhane
Jan 5, 2026 303 Views
Abstract
Co-immunoprecipitation or pull-down assays are frequently used to analyze protein–protein interactions. In these experiments, western blotting is commonly used to detect prey proteins. However, sensitivity and quantification problems remain in this detection system. Recently, the HiBiT-tag-dependent NanoLuc luciferase system was developed as a highly sensitive detection system for small amounts of proteins. In this report, we introduce the method of using HiBiT technology for the detection of prey protein in a pull-down assay. Using this protocol, we demonstrate the formation of a ternary complex consisting of Japanese encephalitis virus NS4B and two host factors, namely valosin-containing protein, and nuclear protein localization protein 4, which is a critical biological event during flavivirus replication in cells.
Background
Most proteins interact with their partner proteins to carry out their biological activity. To understand the role of proteins in cells, co-immunoprecipitation (co-IP) or pull-down assays are frequently used to characterize protein–protein interactions. Western blotting (WB) is commonly used to detect prey proteins in co-IP or pull-down assays. However, its sensitivity is low or dependent on the primary antibody, and it gives inaccurate quantification.
High Bit peptide of NanoLuc Binary Technology (NanoBiT) (HiBiT-tag, amino acid sequence: VSGWRLFKKIS) is a part of split NanoLuc luciferase that can reconstitute intact NanoLuc luciferase when another part of split NanoLuc luciferase, Large Bit peptide of NanoBiT (LgBiT), is present. Therefore, the addition of recombinant LgBiT protein and NanoLuc luciferase substrate and measurement of luminescence facilitates the detection of HiBiT-tag (Dixon et al., 2016). Since HiBiT-tag is a short peptide tag with a length of 11 amino acids, it has a minimum effect on the function of fused proteins. Furthermore, a HiBiT-tag-dependent NanoLuc luciferase detection system is useful for quantifying small quantities of protein, because the signal-to-noise ratio is significantly high in NanoLuc luciferase–dependent luminescence, which allows for small-scale experiments. The procedure is simple and therefore suitable for high-throughput assays. In this report, we introduce the method of using the HiBiT-tag-dependent NanoLuc luciferase system for the detection of prey protein in a pull-down assay. Generally, proteins form an oligomeric complex with various partners. In these cases, the depletion of key subunits significantly affects the formation of the entire complex. In this report, we demonstrate that depletion of a mediator protein affects ternary complex formation.
Japanese encephalitis virus (JEV), a single-stranded positive-sense RNA virus, is a human pathogenic flavivirus. In JEV-infected cells, endoplasmic reticulum membrane–derived large compartments (also called viral replication organelles) are observed. Viruses have been considered to efficiently replicate in these compartments, which may be a target for the development of antiviral reagents (Arakawa and Morita, 2019). Previously, our group reported that the host factor valosin-containing protein (VCP) is recruited to the viral replication organelle and helps in viral genome replication (Tabata et al., 2021). However, no direct interaction between VCP and viral proteins were detected. Our previous study revealed a mediator that bridges the interaction between them. We found that the interaction of nuclear protein localization protein 4 (NPL4), a VCP-associating co-factor, and NS4B, a nonstructural viral protein that localizes on the viral replication site, is important for the recruitment of VCP to the viral replication organelle. We have shown that depletion of NPL4 via siRNA knockdown significantly reduces the affinity between VCP and NS4B, through pull-down assay utilizing HiBiT-tag-dependent luciferase activity (HiBiT activity) measurement (Arakawa et al., 2022) (Figure 1A). HiBiT-tagged NS4B and One-Strep-FLAG (OSF)-tagged VCP were co-expressed in 293T cells and the VCP were affinity-purified using Strep-Tactin beads (Figure 1B). The amount of NS4B in the VCP-bound fraction was then determined in the presence of NPL4 and compared with that in the absence of NPL4 by measuring HiBiT activity (Figure 2C). This protocol is not only useful for studying virus–host interactions, but also has broader applications in the investigation of general protein–protein interactions.
Materials and Reagents
1.5 mL tubes (WATSON, catalog number: 8064131815C)
0.22 μm filter (AS ONE, catalog number: 033022SO-SFCA)
6 cm cell culture dish (Thermo Scientific, catalog number: 150462)
Cell scraper (VIOLAMO, catalog number: 1-2249-01)
White 384-well immuno plates (Thermo Scientific, catalog number: 460372)
293T cells (ATCC: CRL-3216)
Plasmid pCAG-NS4B-FLAG-HiBiT, which encodes HiBiT-tagged JEV NS4B protein (Arakawa et al., 2022)
Plasmid pCAG-OSF-VCP, which encodes One Strep–tagged VCP proteins or is empty (Arakawa et al., 2022)
siRNA-luciferase, which targets firefly luciferase (sense sequence: 3′-CGUACGCGGAAUACUUCGAtt-5′)
siRNA-NPL4, which targets NPL4 (sense sequence: 3′-CUGAAGUGGUCGAUGAAAUtt-5’)
Nano Glo HiBiT lytic detection system (Promega, catalog number: N3040)
Dulbecco’s modified Eagle’s medium (Nacalai Tesque Inc., catalog number: 08458-16)
Fetal bovine serum (FBS) (Thermo Scientific, catalog number: 10270-106)
Phosphate-buffered saline (PBS) without calcium and magnesium (Nacalai Tesque Inc., catalog number: 14249-24)
Benzylpenicillin potassium (Fujifilm Wako Pure Chemical Corporation, catalog number: 021-07732)
Streptomycin sulfate (Tokyo Chemical Industry, catalog number: S0585)
Strep-Tactin Sepharose 50% suspension (IBA Lifesciences GmbH, catalog number: 2-121-010)
Strep-tag elution (10× buffer E) (IBA Lifesciences GmbH, catalog number: 2-1000-025)
cOmplete, EDTA-free, protease inhibitor cocktail (Roche, catalog number: 11873580001)
LipofectamineTM 3000 transfection reagent (Thermo Scientific, catalog number: L3000015)
LipofectamineTM RNAiMAX transfection reagent (Thermo Scientific, catalog number: 13778150)
Opti-MEM (Thermo Scientific, catalog number: 31985062)
Tris (Tris[hydroxymethyl]aminomethane) (Nacalai Tesque Inc., catalog number:35406-91)
NaCl (Nacalai Tesque Inc., catalog number: 31320-05)
Triton X-100 (Nacalai Tesque Inc., catalog number: 35501-15)
100× penicillin G + streptomycin stock solution (see Recipes)
Culture medium (see Recipes)
Pull-down washing buffer (see Recipes)
100× concentrated cOmplete stock solution (see Recipes)
Lysis buffer (see Recipes)
HiBiT reagent (see Recipes)
1× Strep-tag elution buffer (see Recipes)
Equipment
Humidified incubator (37 °C, 5% CO2)
Microplate luminometer (Thermo Scientific, Varioskan LUX Multimode Microplate Reader)
Vortex mixer (Scientific Industries, model: Vortex-Genie 2)
Centrifuge machine for microtube (Thermo Scientific, model: SorvallTM LegendTM Micro 21R)
Rotator (BIO CRAFT, model: BC-710I)
Procedure
Add 60 pmol siRNA-NPL4 (or siRNA-luciferase for negative control) to 250 µL of Opti-MEM. Mix well using a vortex mixer (Figure 1).
Note: Steps 1–8 pertain specifically to the siRNA transfection protocol, while step 9 marks the beginning of the pull-down protocol. To perform siRNA transfection, please follow the "Reverse transfection protocol" of LipofectamineTM RNAiMAX transfection reagent.
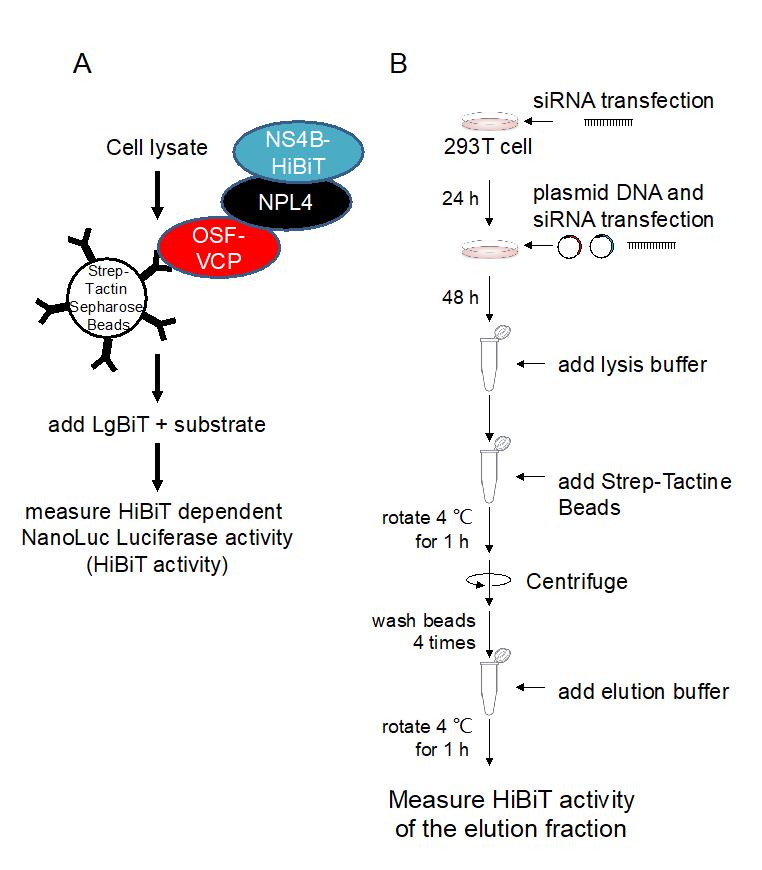
Figure 1. Overview of the protein pull-down HiBiT assay. A. Schematic illustration of the protein pull-down HiBiT assay. B. Flow diagram of the protein pull-down HiBiT assay.Add 6 µL of LipofectamineTM RNAiMAX transfection reagent to 250 µL of Opti-MEM. Mix well using a vortex mixer.
Add the mixture from step 1 to that from step 2 and mix well using a vortex mixer.
Incubate the mixture for 5 min at room temperature.
Seed 293T cells in a 6 cm dish at a density of 750,000 cells/well in 4.5 mL of culture medium.
Gently add approximately 500 µL of the siRNA-Lipofectamine mixture to the cell-containing dish.
Place the dish in a CO2 incubator for 24 h.
Replenish culture medium at 24 h post transfection.
Note: Slowly add medium to the side of the well to avoid cell detachment.
Add 1.5 µg pCAG-NS4B-FLAG-HiBiT, 1.5 µg pCAG-OSF-VCP, and 120 pmol siRNA-NPL4 (or siRNA-luciferase for negative control) to 250 µL of Opti-MEM. For siRNA transfection, repeat the initial transfection using the same siRNA. Mix well using a vortex mixer.
(Optional) Set control experiments using an empty vector (pCAG-OSF) in place of pCAG-OSF-VCP.
Add 6 µL of LipofectamineTM 3000 transfection reagent to 250 µL of Opti-MEM. Mix well using a vortex mixer.
Add the mixture from step 9 to that from step 10 and mix well using a vortex mixer.
Incubate the mixture for 10 min at room temperature.
Apply the mixture from step 12 to the cell-containing dish (~500 µL).
Place the dish in a CO2 incubator for 48 h.
Aspirate the culture medium and add 1 mL of ice-cold PBS to each dish. Collect the cells using a cell scraper and transfer them to a 1.5 mL tube.
Centrifuge the 1.5 mL tubes at 500 × g for 10 min at 4 °C.
Aspirate PBS. Add 1 mL of ice-cold PBS to each tube and suspend by pipetting.
Centrifuge the 1.5 mL tubes at 500 × g for 10 min at 4 °C.
Aspirate PBS. Add 500 µL of ice-cold lysis buffer to each tube and mix by pipetting.
Centrifuge the 1.5 mL tubes at 20,000 × g for 10 min at 4 °C.
Transfer 5 µL of the supernatant to 1.5 mL tubes for input and add 45 µL of lysis buffer. Mix well using a vortex mixer.
Transfer 10 μL of diluted lysate in each well of a 384-well plate.
Note: To ensure accurate measurements, it is recommended to use more than three wells per sample. If the obtained values are outside the reading range, a dilution series should be carried out.
Add 10 μL of HiBiT reagent to each well.
Incubate for 10 min at room temperature.
Measure the luminescence for 1,000 ms measurement time/well using a microplate reader.
Transfer 480 µL of the clear lysate to fresh 1.5 mL tubes.
Add 15 µL of Strep-Tactin Sepharose 50% suspension to the lysate and rotate at 4 °C for 1 h.
Centrifuge the 1.5 mL tubes at 20,000 × g for 1 min at 4 °C.
Aspirate the supernatant. Add 1 mL of cold pull-down washing buffer to each tube and mix via inversion.
Repeat steps 28 and 29 thrice.
Centrifuge the 1.5 mL tubes at 20,000 × g for 1 min at 4 °C.
Aspirate the supernatant. Add 450 µL of 1× Strep-tag elution and rotate at 4 °C for 1 h.
Centrifuge the 1.5 mL tubes at 20,000 × g for 1 min at 4 °C.
Transfer 400 µL of elution buffer to fresh 1.5 mL tubes.
Transfer 10 μL of elution in each well of a 384-well plate.
Note: To ensure accurate measurements, it is recommended to use more than three wells per sample. If the obtained values are outside the reading range, a dilution series should be carried out.
Add 10 μL of HiBiT reagent from Nano Glo HiBiT lytic detection system to each well.
Incubate for 10 min at room temperature.
Measure the luminescence for 1,000 ms measurement time/well using a microplate reader (Figure 2).
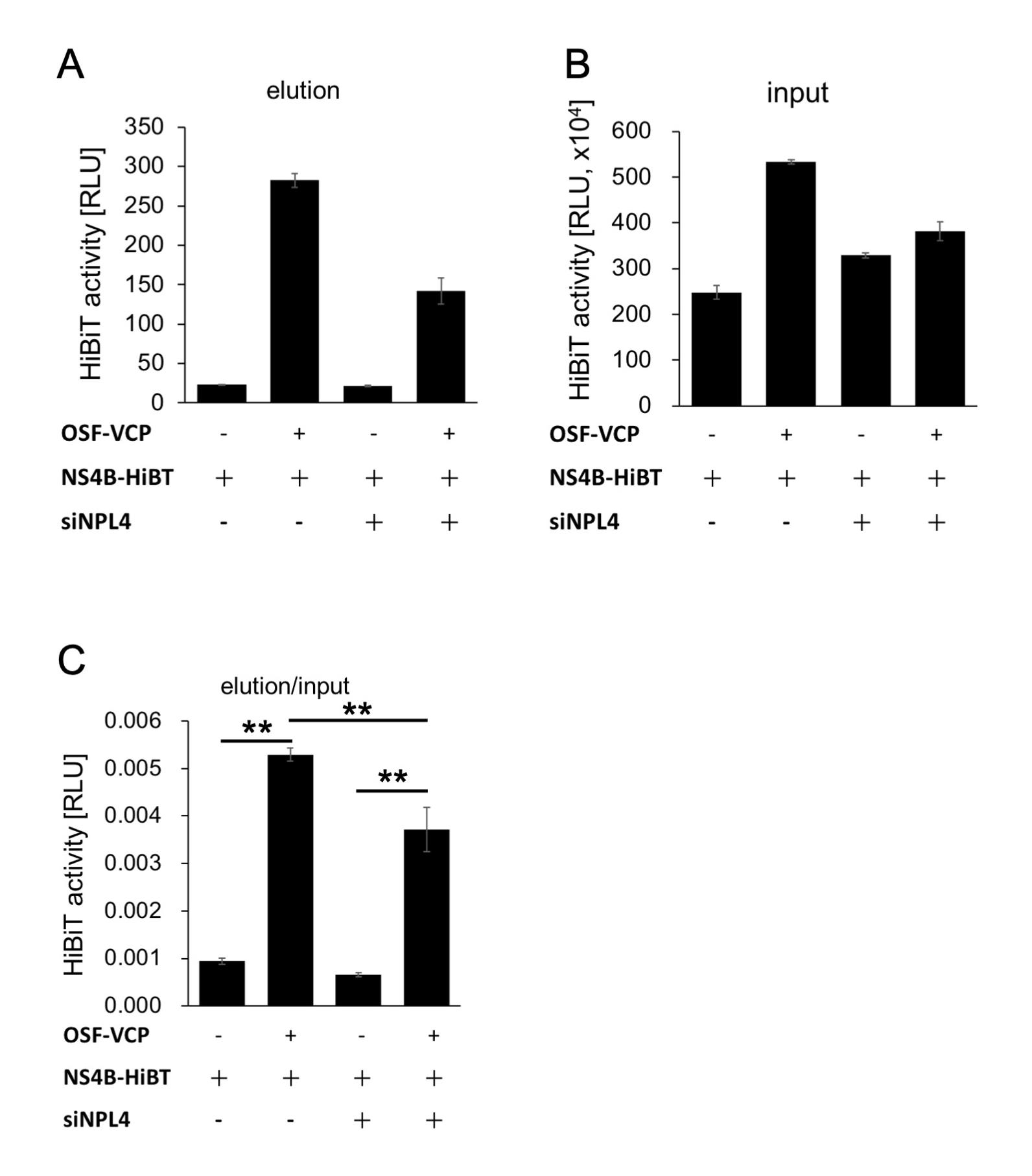
Figure 2. HiBiT activity of the Strep-Tactin-purified fraction and the input fraction. A. HiBiT activity of the Strep-Tactin-purified fraction. B. HiBiT activity of the input fraction. C. The compensated HiBiT value of the Strep-Tactin purified fraction. The HiBiT activity of the Strep-Tactin purified fraction was calculated by dividing its value by that of the input fraction. The means between two groups were compared using a Student’s t-test. Differences were considered significant at **P < 0.01.
Notes
In the NanoLuc/HiBiT system, strong luminescence emission may be obtained and causes leaks to adjacent wells in a multi-well plate. Therefore, we recommend loading the samples in every other well.
The materials used in this study can be obtained from the corresponding author, Eiji Morita, upon reasonable request.
Recipes
100× penicillin G + streptomycin stock solution
PBS 100 mL
Benzylpenicillin potassium 0.626 g
Streptomycin sulfate 1 g
Sterilize using a 0.22 μm filter and store in a refrigerator.
Culture medium
Dulbecco’s modified Eagle’s medium 500 mL
FBS, heat-inactivated via incubation at 56 °C for 45 min 50 mL
100× penicillin G + streptomycin stock solution 5 mL
Pull-down washing buffer
1 M Tris pH 7.5 20 mL
5 M NaCl 30 mL
Triton X-100 1.07 g
ddH2O up to 1 L
100×-concentrated cOmplete stock solution
cOmplete, EDTA-free, protease inhibitor cocktail 1 tablet
ddH2O 500 µL
Store at -20
Lysis buffer
1 M Tris pH 7.5 20 mL
5 M NaCl 30 mL
Triton X-100 10.7 g
ddH2O up to 1 L
Add 100× concentrated cOmplete stock solution just before use.
HiBiT reagent from Nano Glo HiBiT lytic detection system
Nano-Glo HiBiT lytic buffer 500 µl
Nano-Glo HiBiT lytic substrate 10 µl
LgBiT protein 5 µl
1× Strep-tag elution buffer
Strep-tag elution (10× buffer E) 2 mL
ddH2O 18 mL
Acknowledgments
This work was supported by JSPS KAKENHI (Grant Number 22H00553, 22H02873, 22K18378, 20H05305, 20K21874), JST CREST, Japan (grant number JPMJCR17H4). The protocol for HiBiT detection was adapted from previous work (Goto et al., 2020). This protocol is derived from the original research paper (Arakawa et al., 2022; DOI: 10.1016/j.jbc.2022.101597).
Competing interests
The authors have no competing interests directly relevant to the content of this article.
References
- Arakawa, M. and Morita, E. (2019). Flavivirus Replication Organelle Biogenesis in the Endoplasmic Reticulum: Comparison with Other Single-Stranded Positive-Sense RNA Viruses. Int J Mol Sci 20(9).
- Arakawa, M., Tabata, K., Ishida, K., Kobayashi, M., Arai, A., Ishikawa, T., Suzuki, R., Takeuchi, H., Tripathi, L. P., Mizuguchi, K., et al. (2022). Flavivirus recruits the valosin-containing protein-NPL4 complex to induce stress granule disassembly for efficient viral genome replication. J Biol Chem 298(3): 101597.
- Dixon, A. S., Schwinn, M. K., Hall, M. P., Zimmerman, K., Otto, P., Lubben, T. H., Butler, B. L., Binkowski, B. F., Machleidt, T., Kirkland, T. A., et al. (2016). NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem Biol 11(2): 400-408.
- Tabata, K., Arakawa, M., Ishida, K., Kobayashi, M., Nara, A., Sugimoto, T., Okada, T., Mori, K. and Morita, E. (2021). Endoplasmic Reticulum-Associated Degradation Controls Virus Protein Homeostasis, Which Is Required for Flavivirus Propagation. J Virol 95(15): e0223420.
- Goto, S., Ishida, K., Suzuki, R. and Morita, E. (2020). Split Nano Luciferase-based Assay to Measure Assembly of Japanese Encephalitis Virus. Bio Protoc 10(9): e3606.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Arakawa, M. and Morita, E. (2023). Protein Pull-down Assay Using HiBiT-tag-dependent Luciferase Activity Measurement. Bio-protocol 13(6): e4640. DOI: 10.21769/BioProtoc.4640.
- Arakawa, M., Tabata, K., Ishida, K., Kobayashi, M., Arai, A., Ishikawa, T., Suzuki, R., Takeuchi, H., Tripathi, L. P., Mizuguchi, K., et al. (2022). Flavivirus recruits the valosin-containing protein-NPL4 complex to induce stress granule disassembly for efficient viral genome replication. J Biol Chem 298(3): 101597.
Category
Biochemistry > Protein > Interaction > Protein-protein interaction
Microbiology > Microbe-host interactions > Virus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








