- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Separating Inner and Outer Membranes of Escherichia coli by EDTA-free Sucrose Gradient Centrifugation
Published: Vol 13, Iss 6, Mar 20, 2023 DOI: 10.21769/BioProtoc.4638 Views: 2916
Reviewed by: David PaulJose Antonio Reyes-DariasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1790 Views

Mycobacterium smegmatis Ribosome Purification, Co-sedimentation, and Subunit Association Assay
Aneek Banerjee [...] Jayati Sengupta
May 20, 2025 1581 Views

Preparation and Negative Staining for Visualization of Cyanoglobule Lipid Droplets Using Transmission Electron Microscopy
Febri A. Susanto [...] Peter K. Lundquist
Dec 5, 2025 1311 Views
Abstract
The envelope of Gram-negative bacteria consists of an outer membrane (OM), a peptidoglycan cell wall, and an inner membrane (IM). The OM and IM have different components of proteins and lipids. Separating the IM and OM is a basic biochemical procedure to further study lipids and membrane proteins in different locations. Sucrose gradient ultracentrifugation of lysozyme/EDTA-treated total membrane is the most widely used method to separate the IM and OM of Gram-negative bacteria. However, EDTA is often harmful to protein structure and function. Here, we describe a relatively simple sucrose gradient ultracentrifugation method to separate the IM and OM of Escherichia coli. In this method, the cells are broken by a high-pressure microfluidizer, and the total cell membrane is collected by ultracentrifugation. The IM and OM are then separated on a sucrose gradient. Because EDTA is not used, this method is beneficial for subsequent membrane protein purification and functional study.
Background
Gram-negative bacteria are characterized by three layers of cell envelope: an outer membrane (OM), a peptidoglycan cell wall, and an inner membrane (IM) (Silhavy et al., 2010). The IM is a phospholipid bilayer consisting of glycerophospholipids like most plasma membranes, while the OM has an asymmetric lipid structure, with phospholipids in the periplasmic leaflet and lipopolysaccharides (LPS) on the cell surface (Funahara and Nikaido, 1980). Besides the difference in lipid components, the IM and OM also have different membrane protein components (Silhavy et al., 2010). The distinct components bestow their specialized functions: the IM plays a major role in active transport, while the OM protects the cell against harmful compounds like enzymes and large antibiotics and also prevents the leaking of proteins from periplasmic space (DiRienzo et al., 1978).
Separating the IM and OM helps to decipher the location and functions of proteins and lipids in the cell (Schnaitman, 1970) and can also provide the first step in membrane protein purification using styrene-maleic acid (Shu and Mi, 2022). Sucrose gradient ultracentrifugation is the most widely used method to separate the IM and OM of Gram-negative bacteria (Miura and Mizushima, 1968; Osborn et al., 1972a and 1972b; Cian et al., 2020). Lysozyme/EDTA (ethylenediaminetetraacetic acid) is used in those methods to treat the bacterial cells, in which EDTA sequesters divalent cations, thus disrupting the lateral electrostatic bridging interactions between adjacent LPS molecules (Nikaido, 1976). However, the chelating of cations by EDTA may affect the functions or structures of some membrane proteins, which need divalent cations, such as Ca2+, Mg2+, or Zn2+, to stabilize tertiary structure or/and keep activities. Here, we describe a relatively simple sucrose gradient ultracentrifugation method to separate the IM and OM of Escherichia coli, the extensively studied model organism of Gram-negative bacteria. In this method, lysozyme or EDTA are not used, so it is beneficial for subsequent membrane protein purification. To further test the purity of separated OM and IM, anti-OM (e.g., OmpC) and anti-IM proteins (e.g., AcrB) western blot can be used (Guzman-Flores et al., 2017).
Materials and Reagents
125 mL flask (PYREX, catalog number: 4980)
2,800 mL flask (PYREX, catalog number: 4420)
Pipette tips
Serological pipets
1 L centrifuge bottle (Thermo Scientific, NalgeneTM, catalog number: 3141-1006)
70 mL ultracentrifuge bottle (Beckman Coulter, catalog number: 355622)
37 mL ultracentrifuge tube (Beckman Coulter, catalog number: 344058)
Tissue grinder (DWK Life Sciences, catalog number: 8853030040)
Luria broth (Research Products International, catalog number: L24400-5000.0)
Terrific broth (Research Products International, catalog number: T15100-5000.0)
Agar (BD, BactoTM, catalog number: 214010)
Tris (American Bio, catalog number: AB02000-05000)
Sodium chloride (NaCl) (Dot Scientific, catalog number: DSS23020-5000)
Glycerol (J.T. Baker, catalog number: 2136-03)
Kanamycin sulfate (American Bio, catalog number: AB01100-00010)
Spectinomycin (Research Products International, catalog number: S23000-25.0)
Chloramphenicol (Sigma, catalog number: C0378-25G)
Sucrose (Sigma-Aldrich, catalog number: S0389-1 KG)
IPTG (isopropyl β-d-1-thiogalactopyranoside) (American Bio, catalog number: AB00841-00010)
PMSF (phenylmethylsulfonyl fluoride) (Dot Scientific, catalog number: DSP20270-25)
2-Mercaptoethanol (Sigma-Aldrich, catalog number: M6250-100ML)
Glycerol stock of BL21 (DE3) star plysS strain expressing the YejM/LapB complex (Shu and Mi, 2022)
Hydrochloric acid (J.T. Baker, catalog number: 9535-00)
LB medium (see Recipes)
LB agar plate (see Recipes)
TB medium (see Recipes)
1 M Tris-HCl, pH 7.8 (see Recipes)
Lysis buffer (see Recipes)
Dilution buffer (see Recipes)
73% sucrose (see Recipes)
53% sucrose (see Recipes)
20% sucrose (see Recipes)
Equipment
Pipettes (P200, P1000)
Pipette controller (BrandTech Scientific, model: accu-jet pro)
-80 °C ultra-low temperature freezer (PHCbi, model: MDF-U76VA-PA)
Microbiological incubator (VWR, model: F Air 6.3 CF)
Incubator shaker (New Brunswick, model: InnovaTM 44)
Centrifuge (Thermo Scientific Sorvall, model: BP8)
Ultracentrifuge (Beckman, model: OptimaTM LE-80K)
High-pressure microfluidizer (Avestin, model: Emulsi Flex-C3)
Fixed angle rotor (Beckman Coulter, model: Type 45Ti)
Swinging-bucket rotor (Beckman Coulter, model: SW 28)
Ultrapure water system (Millipore, model: Reference A+)
pH meter (Apera, model: pH700)
Procedure
Prepare total cell membrane
Streak the glycerol stock of BL21 (DE3) star plysS strain expressing the YejM/LapB complex on an LB agar plate containing 50 µg/mL of kanamycin sulfate, 50 µg/mL of spectinomycin, and 25 µg/mL chloramphenicol (Kan+Spe+Cam+). Incubate the plate at 37 °C on a microbiological incubator overnight.
Inoculate a single colony from the overnight culturing plate into 20 mL of LB medium (Kan+Spe+Cam+) in a 125 mL flask. Grow the cells at 37 °C with shaking at 220 rpm overnight.
Transfer 10 mL of overnight culture to 1 L of TB medium (Kan+Spe+Cam+) in 2,800 mL flasks (2 L in total). Grow the cells at 37 °C with shaking at 220 rpm to OD600 of 0.6.
Add 200 µL of 1 M IPTG to 1 L of cell culture (final concentration 0.2 mM) and incubate the cells at 30 °C with shaking at 220 rpm for another 4 h.
Centrifuge the culture at 5,000 × g for 10 min at 4 °C to harvest the cells.
Resuspend the cell pellet in 40 mL of lysis buffer (see Recipes) supplemented with 1 mM PMSF and 5 mM 2-Mercaptoethanol. At this step, resuspended cells can be flash-frozen in liquid nitrogen and stored in a -80 °C ultra-low temperature freezer, or move on directly to step A7 to break the cells.
Transfer the cells to a 40 mL tissue grinder. Homogenize the cells by pushing and pulling the grind pestle 10–20 times.
Break the cells in a high-pressure microfluidizer at 15,000 psi with two passes.
Transfer the broken cells to 50 mL centrifuge tubes and remove cell debris by centrifugation at 5,000 × g for 10 min at 4 °C.
Transfer the supernatant to a 70 mL ultracentrifuge bottle and add lysis buffer to the final volume of ~50 mL. Carefully balance the ultracentrifuge bottles.
Ultracentrifuge in a Beckman Coulter type 45Ti rotor at 40,000 rpm (185,677 × g) for 2 h at 4 °C. Keep the membrane pellet after centrifugation.
Separate inner and outer membranes
Resuspend ~3 g of membrane in 55 mL of 20% sucrose with a 40 mL tissue grinder.
Prepare six 37 mL ultracentrifuge tubes. In each tube, from the bottom to the top, layer 14 mL of 73% sucrose, 14 mL of 53% sucrose, and 9 mL of membrane resuspended in 20% sucrose.
Ultracentrifuge without braking at 23,000 rpm (65,000 × g) in a SW 28 rotor for 18 h at 4 °C.
After separation, there should be two bands: the upper band is the IM and the lower one is the OM (Figure 1).
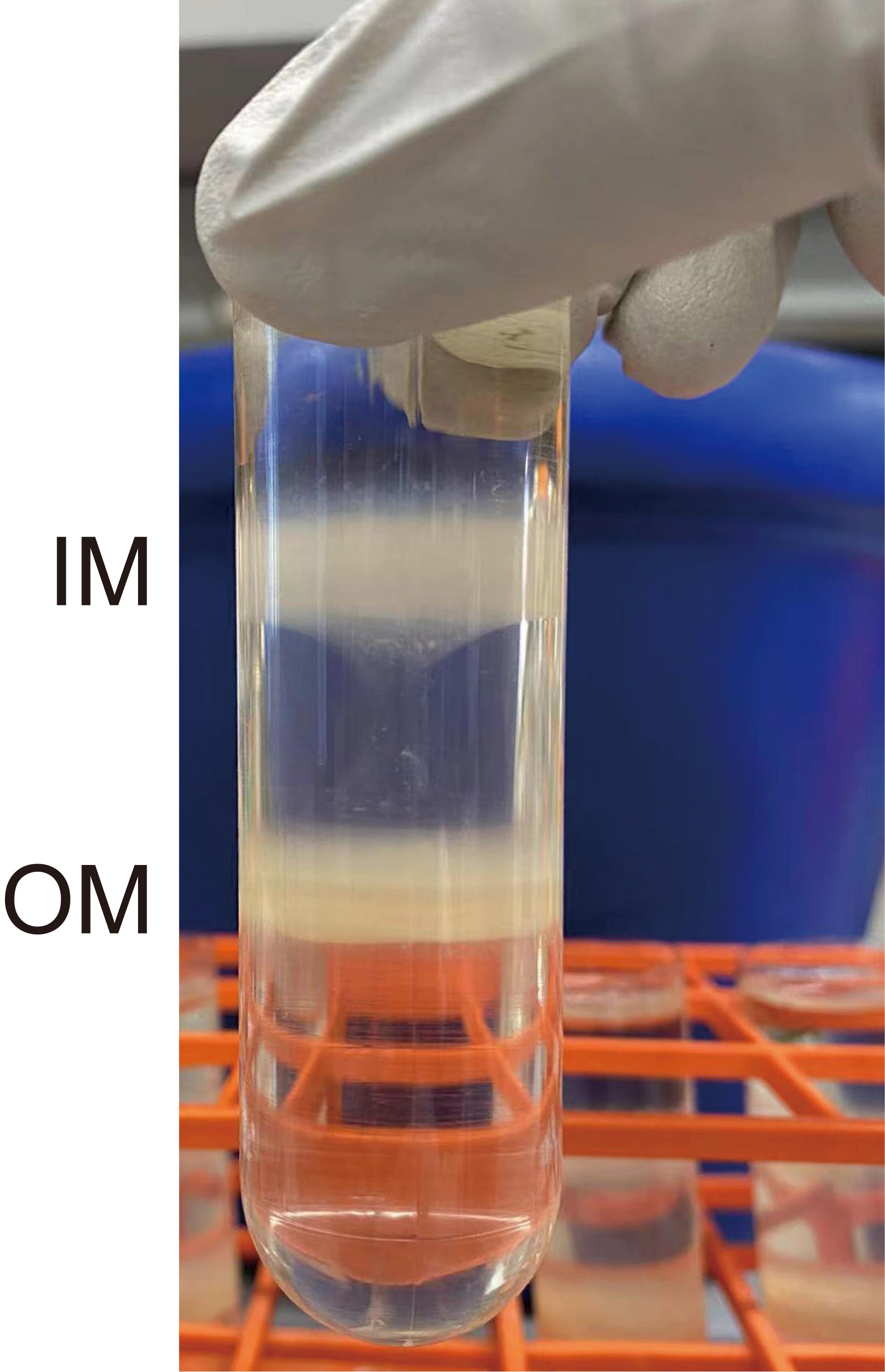
Figure 1. Separation of IM and OMCut the end of a P1000 pipette tip, approximately 5 mm from the point. Collect the upper IM layer using the pipette.
Use a new pipette tip and repeat step B5 to collect the OM layer.
To remove sucrose from the membrane, transfer the IM and OM fractions into 70 mL ultracentrifuge bottles. Add dilution buffer into the bottles to a final sucrose concentration of <10%. Carefully balance the ultracentrifuge bottles.
Ultracentrifuge in a Beckman Coulter type 45Ti rotor at 40,000 rpm (185,677 × g) for 2 h at 4 °C.
Discard the supernatant and weigh the wet weight of the membrane. For 1 g of the membrane, add 10 mL of lysis buffer to resuspend using a 40 mL tissue grinder.
Flash-freeze the membrane with liquid nitrogen and store in a -80 °C ultra-low temperature freezer.
Notes
This protocol uses E. coli strain BL21 (DE3) star plysS expressing the YejM/LapB complex as an example to separate IM and OM. This protocol also applies to other E. coli strains.
All the buffers used in this protocol are kept at 4 °C.
When separating the IM and OM, for each 37 mL ultracentrifuge tube, do not add more than 0.5 g of the total membrane. Otherwise, the IM and OM may not be separated well.
To remove the sucrose in the membrane by ultracentrifuge, the membrane must be first diluted to a final sucrose concentration of less than 10%.
Recipes
LB medium
25 g Luria broth, dissolve in 1 L of deionized water. Autoclave at 121 °C for 30 min.
LB agar plate
LB medium with 1.5% w/v agar.
TB medium
47.6 g Terrific broth and 4 mL of glycerol. Dissolve in 1 L of deionized water.
Autoclave at 121 °C for 30 min.
1 M Tris-HCl, pH 7.8
Dissolve 121.14 g of Tris in 800 mL of Milli-Q water. Adjust the pH to 7.8 using hydrochloric acid. Then add Milli-Q water to a final volume of 1 L.
Lysis buffer
50 mM Tris-HCl, pH 7.8
300 mM NaCl
10% glycerol (v/v)
Dilution buffer
50 mM Tris-HCl, pH 7.8
300 mM NaCl
73% sucrose
73% w/v sucrose
20 mM Tris-HCl, pH 7.8
53% sucrose
53% w/v sucrose
20 mM Tris-HCl, pH 7.8
20% sucrose
20% w/v sucrose
20 mM Tris-HCl, pH 7.8
Acknowledgments
Research reported in this publication was supported by NIAID and NIGMS of the National Institutes of Health under award numbers R21AI156595 and R01GM137068. This protocol was previously used in our reported results (Shu and Mi, 2022).
Competing interests
S. S and W. M declare no conflicts of interest.
References
- Cian, M. B., Giordano, N. P., Mettlach, J. A., Minor, K. E. and Dalebroux, Z. D. (2020). Separation of the Cell Envelope for Gram-negative Bacteria into Inner and Outer Membrane Fractions with Technical Adjustments for Acinetobacter baumannii. J Vis Exp (158). doi: 10.3791/60517.
- DiRienzo, J. M., Nakamura, K. and Inouye, M. (1978). The outer membrane proteins of Gram-negative bacteria: biosynthesis, assembly, and functions. Annu Rev Biochem 47: 481-532.
- Funahara, Y. and Nikaido, H. (1980). Asymmetric localization of lipopolysaccharides on the outer membrane of Salmonella typhimurium. J Bacteriol 141(3): 1463-1465.
- Guzman-Flores, J. E., Alvarez, A. F., Poggio, S., Gavilanes-Ruiz, M. and Georgellis, D. (2017). Isolation of detergent-resistant membranes (DRMs) from Escherichia coli. Anal Biochem 518: 1-8.
- Miura, T. and Mizushima, S. (1968). Separation by density gradient centrifugation of two types of membranes from spheroplast membrane of Escherichia coli K12. Biochim Biophys Acta 150(1): 159-161.
- Nikaido, H. (1976). Outer Membrane of Salmonella-typhimurium Transmembrane Diffusion of Some Hydrophobic Substances. Biochimica Et Biophysica Acta 433(1): 118-132. 1
- Osborn, M. J., Gander, J. E. and Parisi, E. (1972a). Mechanism of assembly of the outer membrane of Salmonella typhimurium. Site of synthesis of lipopolysaccharide. J Biol Chem 247(12): 3973-3986.
- Osborn, M. J., Gander, J. E., Parisi, E. and Carson, J. (1972b). Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem 247(12): 3962-3972.
- Schnaitman, C. A. (1970). Protein composition of the cell wall and cytoplasmic membrane of Escherichia coli. J Bacteriol 104(2): 890-901.
- Shu, S. and Mi, W. (2022). Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli. Nat Commun 13(1): 4576.
- Silhavy, T. J., Kahne, D. and Walker, S. (2010). The bacterial cell envelope. Cold Spring Harb Perspect Biol 2(5): a000414.
Article Information
Copyright
© 2023 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Shu, S. and Mi, W. (2023). Separating Inner and Outer Membranes of Escherichia coli by EDTA-free Sucrose Gradient Centrifugation. Bio-protocol 13(6): e4638. DOI: 10.21769/BioProtoc.4638.
Category
Microbiology > Microbial biochemistry > Other compound
Microbiology > Microbial cell biology > Organelle isolation
Biochemistry > Lipid > Membrane lipid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









