- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
BONCAT-based Profiling of Nascent Small and Alternative Open Reading Frame-encoded Proteins
(*contributed equally to this work) Published: Vol 13, Iss 1, Jan 5, 2023 DOI: 10.21769/BioProtoc.4585 Views: 2096
Reviewed by: ASWAD KHADILKARThomas Farid MartínezMarie A Brunet

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Protein Level Quantification Across Fluorescence-based Platforms
Hector Romero [...] M. Cristina Cardoso
Oct 5, 2023 2000 Views
Sensitive and Adaptable Turn-On Maturation (ATOM) Fluorescent Biosensors for Detecting Subcellular Localization of Protein Targets in Cells
Harsimranjit Sekhon [...] Stewart N. Loh
Mar 20, 2025 2236 Views
Preparation of Testicular Cells for Immunofluorescence Analysis of Manchette in Elongating Spermatids
Changmin Niu [...] Zhibing Zhang
Jun 20, 2025 2354 Views
Abstract
RIBO-seq and proteogenomics have revealed that mammalian genomes harbor thousands of unannotated small and alternative open reading frames (smORFs, <100 amino acids, and alt-ORFs, >100 amino acids, respectively). Several dozen mammalian smORF-encoded proteins (SEPs) and alt-ORF-encoded proteins (alt-proteins) have been shown to play important biological roles, while the overwhelming majority of smORFs and alt-ORFs remain uncharacterized, particularly at the molecular level. Functional proteomics has the potential to reveal key properties of unannotated SEPs and alt-proteins in high throughput, and an approach to identify SEPs and alt-proteins undergoing regulated synthesis should be of broad utility. Here, we introduce a chemoproteomic pipeline based on bio-orthogonal non-canonical amino acid tagging (BONCAT) (Dieterich et al., 2006) to profile nascent SEPs and alt-proteins in human cells. This approach is able to identify cellular stress-induced and cell-cycle regulated SEPs and alt-proteins in cells.
Graphical abstract
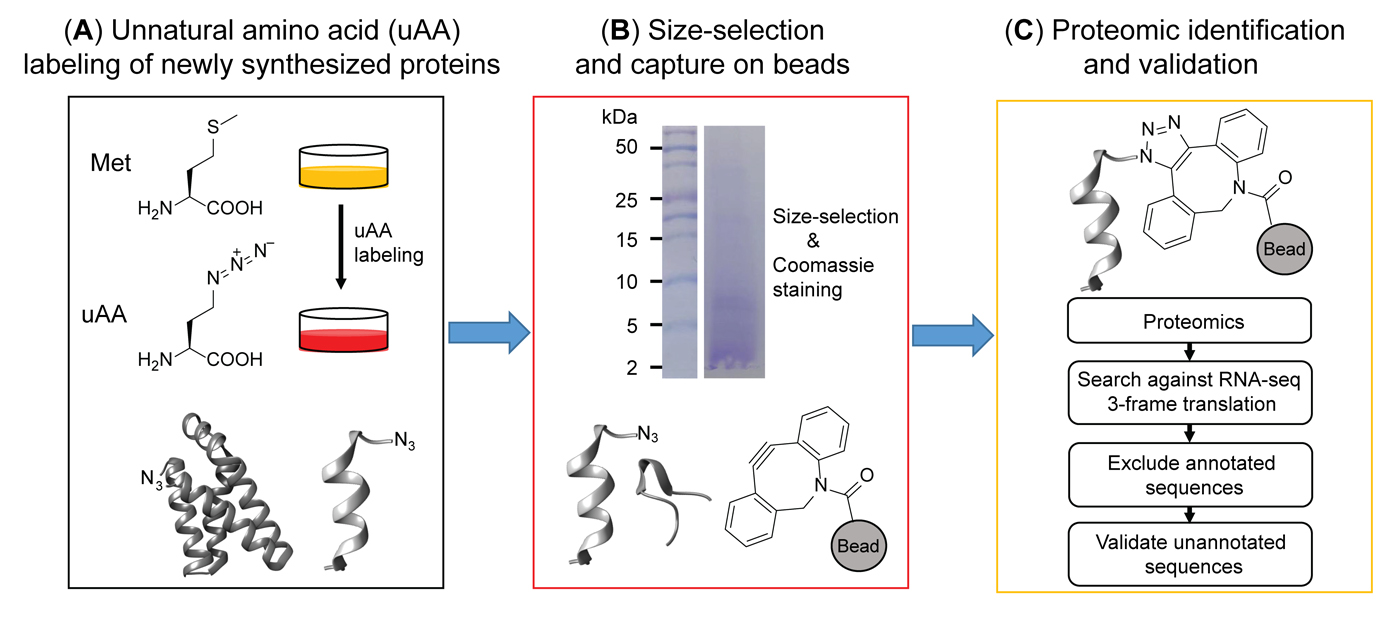
Schematic overview of BONCAT-based chemoproteomic profiling of nascent, unannotated small and alternative open reading frame-encoded proteins (SEPs and alt-proteins)
Background
Thousands of previously unannotated, expressed small open reading frames (smORFs) and alternative open reading frames (alt-ORFs) have recently been identified in mammalian genomes (Orr et al., 2020). These smORFs and alt-ORFs are found in 5' and 3' untranslated regions of mRNAs, in frame-shifted ORFs overlapping protein-coding sequences, and within long noncoding RNAs (including pseudogenes and antisense RNAs) (Brunet et al., 2018). Some smORF-encoded proteins (SEPs, also termed micropeptides or microproteins) and alt-ORF-encoded proteins (alt-proteins), which we will collectively refer to as “small proteins” in this protocol (Orr et al., 2020), have been shown to play important biological roles (Chen et al., 2020; Cao et al., 2021; Magny et al., 2021), suggesting that defining the functions of small proteins represents a major opportunity to gain insights into biology. However, due to their short lengths and limited homology to protein domains of known function, the majority of unannotated small proteins remain uncharacterized.
Similar to annotated proteins, the expression of some unannotated small proteins is cell-type specific (Cao et al., 2020), and is regulated by cellular stress (Jackson et al., 2018; Zhang et al., 2022). However, many prior reports on the discovery of unannotated small proteins do not provide information about differential expression or regulation. Methods to detect small proteins that are regulated by (patho)physiological processes and cellular stress should be of broad utility to enable hypothesis generation about their functions in high throughput.
Bio-orthogonal non-canonical amino acid tagging (BONCAT) incorporates the methionine analog azidohomoalanine (AHA) into all cellular proteins synthesized by the endogenous protein translation machinery during the labeling window (Dieterich et al., 2006). The first reported BONCAT workflows derivatize AHA-labeled proteins with biotin-alkyne, requiring a column-based step to remove excess biotin-alkyne, which de-enriches small proteins prior to streptavidin capture of the labeled proteome (Cao et al., 2022). Here, we describe a modified approach to profile nascent small proteins, which interfaces AHA labeling with in-solution size selection, followed by click chemistry capture directly on dibenzocyclooctyne magnetic beads, enabling sensitive detection of small proteins using mass spectrometry proteomics coupled with bioinformatic methods for unannotated protein identification. In a proof-of-principle study demonstrating this approach, we identified 22 actively translated, unannotated small proteins and N-terminal extensions of canonical proteins, in a cultured human cell line under control and stress conditions; we further confirmed that one of these unannotated small proteins is post-transcriptionally upregulated by DNA damage stress, and another one is cell-cycle-regulated (Cao et al., 2022), suggesting that this method may be broadly useful to reveal the regulated synthesis of unannotated small proteins in cultured cells.
Materials and Reagents
15-cm cell culture dish (Falcon, catalog number: 353025)
6-well cell culture plate (Greiner Bio-One, catalog number: 657160)
15 mL tube (Falcon, catalog number: 352096)
PolyWAX LPTM column (PolyLC, catalog number: 104WX0510)
Dulbecco’s modified Eagle’s medium (DMEM) (Corning, catalog number: 10-013-CV)
DMEM without methionine (DMEM-Met) (Corning, catalog number: 17-204-CI)
L-Azidohomoalanine (AHA) (Click Chemistry Tools, catalog number: 1066-100)
Fetal bovine serum (FBS) (Sigma, catalog number: F4135)
Penicillin/streptomycin (Pen/Strep) (Gibco, catalog number: 15140-122)
Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) (Click Chemistry Tools, catalog number: 1061-100)
Copper(II) sulfate (CuSO4) (Sigma, catalog number: C1297)
Biotin-alkyne (PEG4 carboxamide-Propargyl Biotin) (Click Chemistry Tools, catalog number: 1266-5)
Bond Elut C8 cartridge (Agilent, catalog number: 12105028)
DBCO (dibenzocycloctyne) magnetic beads (Click Chemistry Tools, catalog number: 1037-1)
2-Iodoacetamide (TCI, catalog number: I0741, CAS 144-48-9)
Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (Sigma, catalog number: C4706)
Sequencing grade modified trypsin (Promega, catalog number: V5111)
Ethyl acetate (Sigma, catalog number: 270989)
C18 spin columns (Thermo Scientific, catalog number: 89873)
Triethylamine (TEA) (Sigma, catalog number: 471283)
88%–91% formic acid (FA) (Sigma, catalog number: 399388)
Acetonitrile (ACN) (Sigma, catalog number: 271004)
Trifluoroacetic acid (TFA) (Sigma, catalog number: 302031)
Methanol (Sigma, catalog number: 34860)
Chloroform (Avantor, catalog number: JT-9180-01)
Triton X-100 (Amresco, catalog number: 0694)
DMSO (Dimethyl sulfoxide) (Alfa Aesar, catalog number: 36480)
Streptavidin-HRP (Invitrogen, catalog number: S911)
Ammonium bicarbonate (NH4HCO3) (Sigma, catalog number: A6141)
Urea (CH4N2O) (Sigma, catalog number: U5128)
Potassium chloride (KCl) (Avantor, catalog number: 3040-01)
Sodium carbonate (Na2CO3) (Sigma, catalog number: 230952)
Calcium chloride (CaCl2) (MP Biomedicals, catalog number: 153502)
Lysis buffer (see Recipes)
0.25 M TEAF pH 3.0 (see Recipes)
RIPA buffer (see Recipes)
4× SDS loading buffer (see Recipes)
Water-saturated ethyl acetate (see Recipes)
Equipment
Refrigerated centrifuge (Eppendorf, model: 5424 R)
Refrigerated centrifuge (Eppendorf, model: 5810 R)
SpeedVac vacuum concentrator (Thermo Scientific, Savant SPD10)
Vortex mixer (VWR, G-560 Vortexer 2)
Tube rotator (Thermo Scientific, catalog number: 05-450-127)
Heat block (Thermo Scientific)
37 °C incubator (Thermo Scientific)
Q Exactive Plus Hybrid Quadrupole Orbitrap (Thermo Scientific)
-80 °C freezer (Thermo Scientific)
-20 °C freezer (Thermo Scientific)
NEB magnetic holder (NEB, S1509S)
Agilent 1100 HPLC (Agilent)
Additional reagents and equipment for standard immunoblotting procedures
Software
Thermo Scientific Xcalibur (version 4.4)
Mascot (version 2.5.1)
Image Lab (version 5.2)
Agilent Chemstation for LC systems (version B.04.03)
Procedure
AHA labelling of newly synthesized proteins
Seed 2.5 × 107 HEK 293T cells in a 15-cm cell culture dish, followed by culture in complete DMEM with 10% FBS and 1% Penicillin/Streptomycin in a 5% CO2 atmosphere at 37 °C overnight, to achieve 80%–90% confluency.
Aspirate medium from the cells, wash once with 10 mL of room-temperature (RT) 1× PBS, then add 10 mL of pre-warmed DMEM-Met, and incubate in a 5% CO2 atmosphere at 37 °C for 30 min.
Prepare 0.4 M AHA (100× stock, which is stable at -20 °C for at least 2 months) in 1× PBS. Gently remove DMEM -Met from the cells, then gently add 10 mL of pre-warmed DMEM-Met with 4 mM AHA (1×) and 10% FBS. Incubate in a 5% CO2 atmosphere at 37 °C for 2 h, to label the newly synthesized proteins with AHA.
Remove the medium, wash twice with 10 mL of 1× PBS, and harvest the cells by pipetting up and down (Note: other adherent cell types likely need to be harvested with gentle scraping). Transfer the cells into a 15 mL tube, centrifuge at 800 × g and RT for 3 min, and gently aspirate the supernatant.
Note: Samples can be flash frozen and stored for 1–2 months at -80 °C at this point.
Validation of AHA labelling and C8 column-based size-selection
Especially if performing BONCAT labeling for the first time, it is important to confirm AHA incorporation into the cellular proteome, prior to proceeding to proteomic analysis. To detect AHA-labeled proteins, we recommend that the cells from a single well of a 6-well cell culture plate, with protein concentration-normalized lysate from unlabeled or vehicle-treated cells as a negative control, be subjected to Click chemistry with commercially available biotin-alkyne, followed by streptavidin-HRP blotting, as shown in Figure 1. This step can be performed on a separate sample that has been prepared in parallel to the cells to be used for proteomic analysis.
Add 200 µL of 1% w/v SDS in 1× PBS to the cell pellet, followed with boiling at 100 °C for 10 min. In parallel for all steps, process an unlabeled (non-AHA-treated) pellet of an equal number of the same cells as a negative control, to assess labeling efficiency over background.
Add 1.8 mL of 1× PBS containing 0.2% Triton X-100 to the cell lysate.
For 1 mL of total lysate, prepare a mixture by adding 2 µL of a 350 mM stock of CuSO4 in water, 1 µL of 200 mM TBTA stock in DMSO, and 3.5 µL of 0.5 M TCEP stock in water. Mix and incubate at RT for 10 min, to form the click chemistry catalyst in situ.
Note: The remaining 1 mL of lysate can be reserved for 1–2 days at -20 °C, in case repeat analysis is required; if the remaining lysate is not required, it can be discarded.
Add 1 µL of 25 mM biotin-alkyne in DMSO to the mixture from step B1c, then add this mixture to 1 mL of the cell lysate from step B1b. Incubate with rotation at RT for 6 h, to derivatize AHA-labeled proteins with biotin via click chemistry.
Transfer 100 µL of the click chemistry reaction to a new 1.5-mL tube, add 400 µL of methanol, and vortex for 5 s.
Add 100 µL of chloroform and vortex for 5 s.
Add 300 µL of ddH2O and vortex for 5 s, followed by centrifugation at 18,407 × g at RT for 2 min.
Remove and discard the top layer, add 400 µL of methanol to the remaining bottom layer, vortex for 5 s, and centrifuge at 18,407 × g at RT for 2 min. You should observe the formation of a protein pellet.
Remove the supernatant without disturbing the pellet, and dry it at RT for 5 min. To resuspend the pellet containing labeled cellular proteins, add 50 µL of 1× SDS loading buffer, then boil at 100 °C for 10 min.
Load the resuspended labeled proteins onto a 12% SDS-PAGE gel, followed by transfer to nitrocellulose, and western blotting with streptavidin-HRP, as previously described (Cao et al., 2021). Perform Ponceau staining of the membrane after transfer, to confirm equal loading.
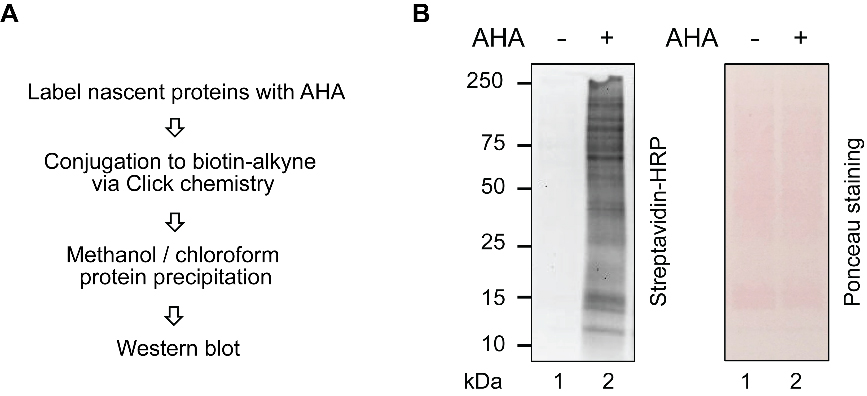
Figure 1. AHA labeling validation with streptavidin-HRP blotting.(A) Experimental scheme for AHA labeling validation. (B) Streptavidin-HRP blotting reflects AHA labeling, and Ponceau staining served as a loading control.
Note: If AHA labeling is inefficient, AHA treatment time and concentration can be optimized.
If the AHA labeling is successful in the replicate sample, add 2 mL of lysis buffer (see Recipes) to an AHA labeled HEK 293T cell pellet from a 15-cm dish from step A4 and vortex, followed by boiling at 100 °C for 10 min. Repeat vortexing and boiling three to four times, until the cell lysate becomes clear. We typically aliquot the lysate into four 1.5-mL Eppendorf tubes for the boiling step, to fit the wells of our heating block, then recombine the lysates for step B3c below, but this can be adapted to any appropriate tube size or boiling apparatus.
Size select to enrich small proteins with two C8 columns, as follows (Ma et al., 2016):
Add 500 µL of methanol to each of the two C8 columns, centrifuge at 200 × g for 1 min, and discard the flow-through.
Add 1 mL of 0.25 M TEAF pH 3.0 (see Recipes) to each of the two C8 columns, then centrifuge at 200 × g for 1 min, and discard the flow-through.
Load 1 mL of the cell lysate from step B2 to each of the two C8 columns (e.g., combined lysates from two Eppendorf tubes for each C8 column), then centrifuge at 200 × g for 2 min, and discard the flow-through.
Add 1 mL of 0.25 M TEAF pH 3.0 to each of the two C8 columns, then centrifuge at 200 × g for 2 min, and discard the flow-through.
Add 0.5 mL of 3:1 ACN:0.25 M TEAF pH 3.0 to each of the two C8 columns, centrifuge at 200 × g for 2 min, and retain the eluate. Repeat this step one more time, and combine all eluates from all columns.
Speedvac the solution to dryness at RT. This step may take up to 3 h.
Click capture on beads
Wash 50 µL of DBCO bead slurry with 1 mL of RIPA buffer (see Recipes), place the tube on a magnetic holder, remove and discard the RIPA buffer.
Add 1.2 mL of RIPA buffer to the size selected cell lysate from step B3f , vortex for 10 s, then resuspend the washed DBCO beads in the cell lysate. Rotate at RT for 1 h, then place the tube on a magnetic holder, remove and discard the supernatant.
Wash the DBCO beads as follows:
Add 1 mL of RIPA buffer to the beads, rotate at RT for 3 min, then place the tube on a magnetic holder, and remove the supernatant.
Add 1 mL of 1 M KCl in ddH2O to the beads, rotate at RT for 3 min, then place the tube on a magnetic holder, and remove the supernatant.
Add 1 mL of 0.1 M Na2CO3 in ddH2O to the beads, rotate at RT for 1 min, then place the tube on a magnetic holder, and remove the supernatant.
Add 1 mL of 2 M urea in ddH2O to the beads, rotate at RT for 1 min, then place the tube on a magnetic holder, and remove the supernatant.
Add 1 mL of RIPA buffer to the beads, rotate at RT for 1 min, then place the tube on a magnetic holder, and remove the supernatant. Repeat this step one more time.
Add 1 mL of 1× PBS to the beads, rotate at RT for 1 min, then place the tube on a magnetic holder, and remove the supernatant. Repeat this step five more times.
On-bead digestion
Add 400 µL of 6 M urea in 1× PBS to the DBCO beads from step C3f , then add 20 µL of 0.2 M TCEP in ddH2O.
Incubate the beads at 60 °C for 10 min, then cool down to RT.
Add 20 µL of 0.4 M 2-iodoacetamide in ddH2O, then incubate with shaking at 37 °C for 15 min.
Dilute the solution by adding 950 µL of 1× PBS, then place the tube on a magnetic holder, and remove the supernatant.
Add 300 µL of a premixed solution of 2 M urea in 1× PBS, with 3 µL of 0.1 M CaCl2 in ddH2O, and 10 µL of 0.5 µg/µL trypsin; incubate in an incubator or water bath at 37 °C for 14–16 h.
Place the tube on a magnetic holder, and transfer the supernatant to a new tube. Wash the beads with 50 µL of water twice to liberate any remaining bound peptides, and combine the washes with the supernatant.
Add 15 µL of 88%–91% FA to stop the digestion.
Speedvac the solution to dryness at RT. This step may take up to 3 h.
Note: Samples can be stored for 1–2 months at -80 °C at this point.
Detergent removal with ethyl acetate
Add 100 µL of 100 mM NH4HCO3 in ddH2O to the digested peptides from step D8 , and vortex for 10 s.
Add 1 mL of water-saturated ethyl acetate (see Recipes) to the digested peptides solution, vortex for 10 s, followed by centrifugation at 18,407 × g at RT for 30 s. Remove and discard the upper layer. Repeat this step one more time. The remaining bottom layer contains the digested peptides.
Speedvac the solution to dryness at RT. This step may take 1 h.
De-salt peptides with a C18 spin column
Add 100 µL of 5% ACN + 0.5% TFA in ddH2O to the digested peptides from step E3, and vortex for 10 s.
Add 200 µL of 50% methanol in ddH2O to a C18 spin column, then centrifuge at 200 × g for 2 min, and discard the flow-through.
Add 150 µL of 5% ACN + 0.5% TFA in ddH2O to the C18 spin column, then centrifuge at 200 × g for 2 min, and discard the flow-through. Repeat this step one more time.
Add the digested peptide solution to the C18 spin column, centrifuge at 200 × g for 2 min, and discard the flow-through.
Wash the C18 spin column with 150 µL 5% ACN + 0.5% TFA in ddH2O, centrifuge at 200 × g for 2 min, and discard the flow-through. Repeat this step one more time.
Elute the digested peptides with 45 µL of 70% ACN in ddH2O, centrifuge at 200 × g for 2 min, and retain the eluate. Repeat this step one more time, and combine the two eluates.
Speedvac the solution to dryness at RT. This step may take up to 2 h.
Note: Samples can be stored for 1–2 months at -80 °C at this point.
Mass spectrometry analysis of nascent small proteins
Optional steps: ERLIC fractionation with a PolyWAX LP column using Agilent 1100 HPLC, as shown in Figure 2.
Add 55 µL of 85% ACN/0.1% FA in ddH2O to the digested peptides from step F7, vortex for 10 s, and load 50 µL of this onto a PolyWAX LP column (150× 1.0-mm; 5 µm 300 Å). Retain 10% of the total peptide suspension for prefractionation LC-MS/MS analysis, as a quality control step.
Separate the peptides with a 80-min gradient, as follows (solvent A: 80% ACN 0.1% FA in ddH2O; solvent B: 30% ACN 0.1% FA in ddH2O): set the flow rate to 300 μL/min, start with 0% B for 5 min, then 0% B to 8% B over 17 min, 8% B to 45% B over 25 min, and 45% B to 100% B over 10 min. Maintain the isocratic flow at 100% B for 5 min, then gradient from 100% B to 0% B over 10 min, and maintain the isocratic flow at 0% B for 8 min.
Collect fractions 1–10 at 1-min intervals, fractions 11–13 at 10-min intervals, and fractions 14–15 at 20-min intervals.
Dry the fractions and the 10% unfractionated peptides in the Speedvac, and re-suspend each sample in 10 µL of 0.1% FA in ddH2O before LC-MS/MS analysis.
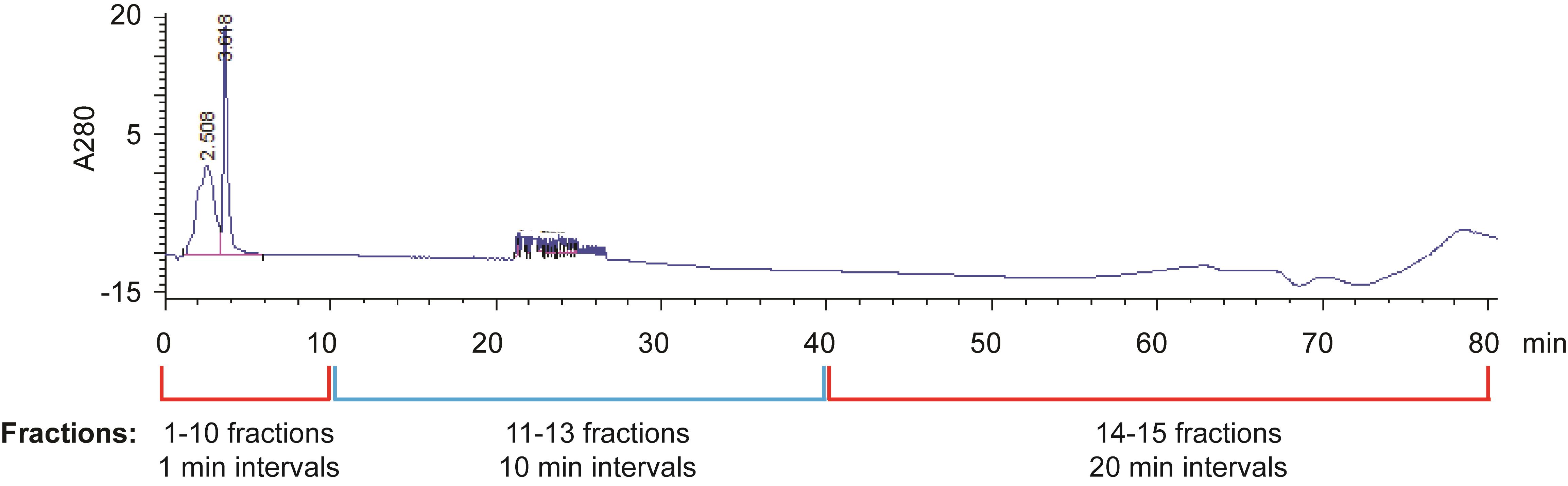
Figure 2. ERLIC fractionation of digested peptidesIf ERLIC fractionation has not been performed, add 35 µL of 0.1% FA in ddH2O to the digested peptides from step F7. Centrifuge at 18,407 × g and 4°C for 30 min, and discard the pellet.
If ERLIC fractionation has been performed, inject 5 µL of each fraction, as well as 5 µL of 10% unfractionated peptides, directly on a prepacked column attached to a nanoAcquity UPLC (Waters) in-line with a Thermo Scientific Q Exactive Plus Hybrid QuadrupoleOrbitrap mass spectrometer. If ERLIC has not been performed, 5 µL of the total digested peptides is injected.
Separate the peptides with a 130-min gradient as follows (solvent A: 0.1% FA in ddH2O; solvent B: ACN with 0.1% FA in ddH2O): set the flow rate to 0.1 μL/min, start with 1% B for 40 min, then gradients 1% B to 6% B over 2 min, 6% B to 24% B over 48 min, 24% B to 48% B over 5 min, and 48% B to 80% B over 5 min. Maintain the isocratic flow at 80% B for 5 min, then gradient from 80% B to 1% B over 5 min, and maintain the isocratic flow at 1% B for 10 min.
Set the mass range to 300–1,700 m/z with a resolution of 70,000, and the automatic gain control (AGC) target to 3 × 106. Collect the MS/MS data using a top 10 high-collisional energy dissociation (HCD) fragmentation method in data-dependent mode, with a normalized collision energy of 27.0 eV and a 1.6 m/z isolation window with a 17,500 MS/MS resolution, and a 90 s dynamic exclusion, according to the manufacturer’s instructions.
Note: Mass spectrometry parameters may require optimization for the instrument available to the experimenter, for example in consultation with a mass spectrometry core facility.
Data analysis
Peptide and protein identification
Convert raw data files to Mascot Generic File (MGF) files, and, for combined annotated and unannotated protein identification, search the data against a database comprised of a three-frame translation of assembled transcripts from RNA-seq data, as previously described (Khitun and Slavoff, 2019), plus a contaminants database provided by Mascot, using Mascot (version 2.5.1). Search the same datasets against the UniProt human proteome database plus a contaminants database separately, for quality control: in a small protein BONCAT experiment without ERLIC fractionation, a typical run detects 1,968 annotated human proteins, and 4.88% are typically <100 amino acids.
Set oxidation of methionine and N-terminal acetylation as variable modifications, and carbamidomethylation of cysteine as a fixed modification. Set enzyme specificity to semiTrypsin, and allow a maximum missed cleavage of 2. Set peptide charge to 2+, 3+ and 4+. Set the peptide mass tolerance to 20 ppm, and fragment mass tolerance to 0.02 Da, as shown in Figure 3. Click Run. The search may take up to 8 h.
Note: We do not set azidohomoalanine as a variable modification during the search, because those fragments containing azidohomoalanine remain bound to the beads, and were removed during digested peptides extraction. We will identify other tryptic peptides from the same protein.
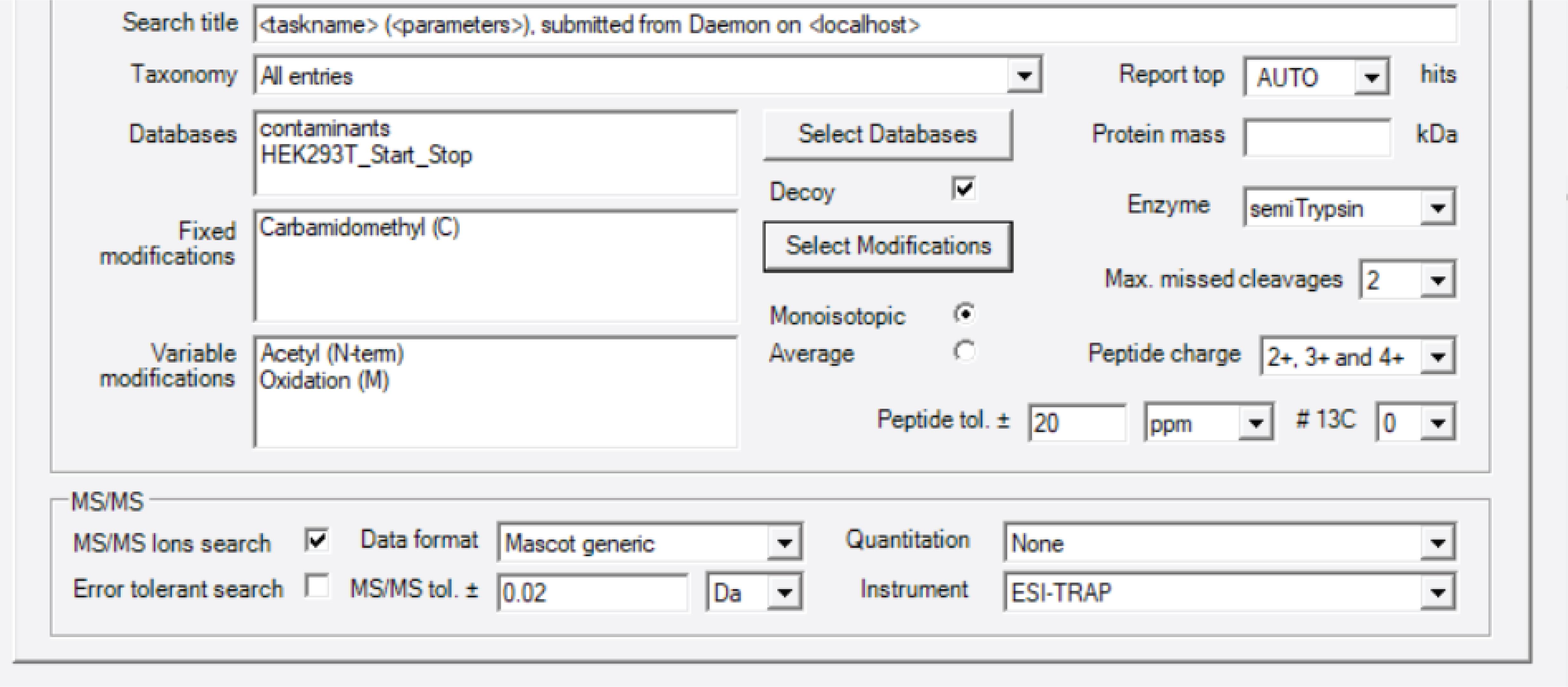
Figure 3. Mascot parameter settingOnce the run is finished, open the Result File URL. Identify and validate the unannotated peptides as shown in our previously published protocol paper (Khitun and Slavoff, 2019). Briefly, peptide-spectral matches to annotated proteins are removed with a custom script available via Zenodo (https://doi.org/10.5281/zenodo.5921116), via matching against a locally downloaded file containing the current human proteome annotation. Remaining peptide-spectral matches are subjected to NCBI protein BLAST against the non-redundant (NR) human protein database, using default parameters for short input sequences to confirm their uniqueness relative to annotated proteins. Any peptides found to be less than two amino acids different from the nearest annotated protein (predicted proteins with NP or XP designation are not included) match are discarded, to eliminate the possibility of false positive matches due to mutations or post-translational modifications. We note that amino acids I and L are isobaric and cannot be distinguished by mass spectrometry, so peptides differing an annotated protein by two amino acids, but one of the amino acids is I or L, should also be discarded. Because small protein identifications often rely on only a single peptide-spectral match, MS/MS spectra are manually inspected for five additional key parameters (Figure 4, right). Peptide-spectral matches that do not pass these criteria are discarded. A high-quality MS/MS spectrum from a recently identified alt-protein called MINAS-60 (Cao et al., 2022) is shown in Figure 4, as an example.
Peptide-spectral matches that pass the filters are computationally mapped to their encoding transcripts and unique genomic loci. Briefly, transcript sequences corresponding to the candidate unannotated peptides identified in step 3 are extracted, which can be found in the transcript database used to generate the 3-frame proteomic search database, and then translated into amino acid sequences in three frames in a format amenable to visual analysis using the ExPASy translate tool. The smORF encoding each unannotated tryptic peptide is identified based on the presence of the tryptic peptide sequence within the translated region corresponding to the smORF. Experimental validation of smORF expression at the molecular level is ultimately required to assign a novel SEP, and exclude the remaining possibility of false positive identifications (for example, resulting from multiple isobaric substitutions within an assigned peptide-spectral match), as previously described (Khitun and Slavoff, 2019).
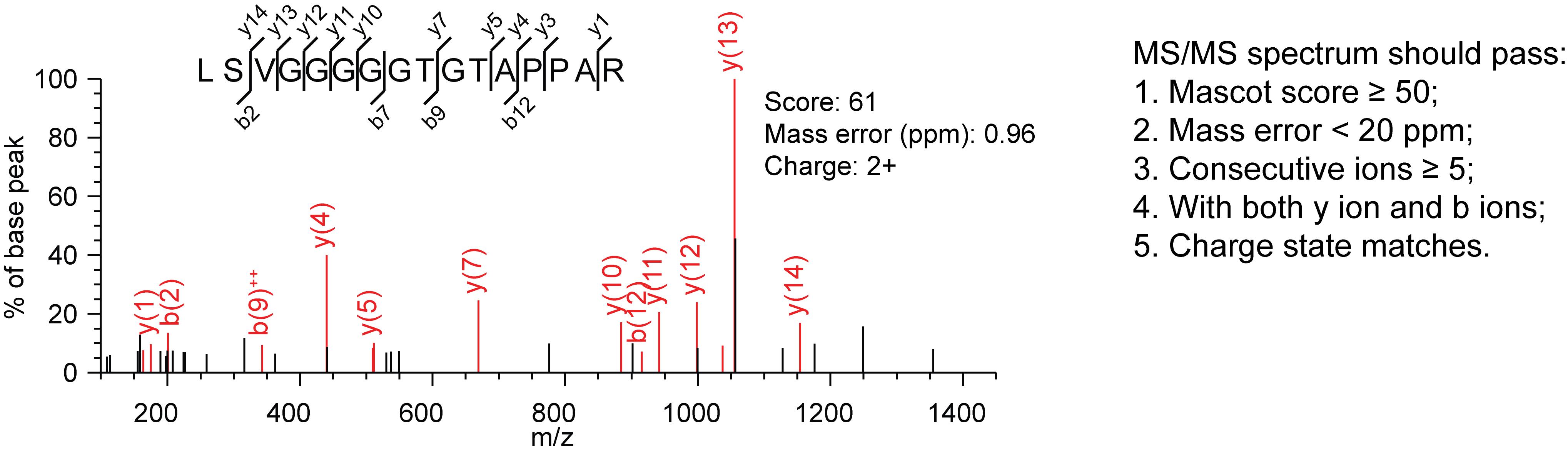
Figure 4. High-quality MS/MS spectrum from MINAS-60. Y and b ions are product ions generated from fragmentation of the parent tryptic peptide. A y ion is an ionized peptide fragment bearing charge on the C-terminus, and a b ion is a peptide fragment ion bearing the charge on the N-terminus.
Recipes
Lysis buffer
50 mM HCl
0.01% β-mercaptoethanol (v/v)
0.05% Triton X-100 (v/v)
0.25 M TEAF pH3.0
To make a 50 mL solution, add 2 mL triethylamine and 2 mL formic acid to 46 mL ddH2O.
RIPA buffer
10 mM Tris-HCl (pH = 7.4)
1% Triton X-100 (v/v)
0.1% sodium deoxycholate (w/v)
0.1% SDS (w/v)
140 mM NaCl
4× SDS loading buffer
240 mM Tris-HCl (pH = 6.8)
40% glycerol (v/v)
8% SDS (w/v)
5% β-mercaptoethanol (v/v)
0.04% bromophenol blue (w/v)
Water-saturated ethyl acetate
To make a 50 mL solution, add 5 mL ddH2O to 45 mL ethyl acetate.
Acknowledgments
This work was supported by a Mark Foundation for Cancer Research Emerging Leader Award (21-055-ELA), a Paul G. Allen Frontiers Group Distinguished Investigator Award, a Sloan Research Fellowship (FG-2022-18417), a Searle Scholars Program Award, an Odyssey Award from the Richard and Susan Smith Family Foundation, and start-up funds from Yale University West Campus (to S. A. S.). X.C. was supported in part by a Rudolph J. Anderson postdoctoral fellowship from Yale University. A.K. was in part supported by an NIH Predoctoral Training Grant (5T32GM06754 3-12).
Competing interests
The authors declare no competing interests.
References
- Brunet, M. A., Levesque, S. A., Hunting, D. J., Cohen, A. A. and Roucou, X. (2018). Recognition of the polycistronic nature of human genes is critical to understanding the genotype-phenotype relationship. Genome Res 28(5): 609-624.
- Cao, X., Khitun, A., Harold, C. M., Bryant, C. J., Zheng, S. J., Baserga, S. J. and Slavoff, S. A. (2022). Nascent alt-protein chemoproteomics reveals a pre-60S assembly checkpoint inhibitor.Nat Chem Biol 18(6): 643-651.
- Cao, X., Khitun, A., Luo, Y., Na, Z., Phoodokmai, T., Sappakhaw, K., Olatunji, E., Uttamapinant, C. and Slavoff, S. A. (2021). Alt-RPL36 downregulates the PI3K-AKT-mTOR signaling pathway by interacting with TMEM24. Nat Commun 12(1): 508.
- Cao, X., Khitun, A., Na, Z., Dumitrescu, D. G., Kubica, M., Olatunji, E. and Slavoff, S. A. (2020). Comparative Proteomic Profiling of Unannotated Microproteins and Alternative Proteins in Human Cell Lines. J Proteome Res 19(8): 3418-3426.
- Chen, J., Brunner, A. D., Cogan, J. Z., Nunez, J. K., Fields, A. P., Adamson, B., Itzhak, D. N., Li, J. Y., Mann, M., Leonetti, M. D., et al. (2020). Pervasive functional translation of noncanonical human open reading frames. Science 367(6482): 1140-1146.
- Dieterich, D. C., Link, A. J., Graumann, J., Tirrell, D. A. and Schuman, E. M. (2006). Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT).Proc Natl Acad Sci U S A 103(25): 9482-9487.
- Jackson, R., Kroehling, L., Khitun, A., Bailis, W., Jarret, A., York, A. G., Khan, O. M., Brewer, J. R., Skadow, M. H., et al. (2018). The translation of non-canonical open reading frames controls mucosal immunity.Nature 564(7736): 434-438.
- Khitun, A. and Slavoff, S. A. (2019). Proteomic Detection and Validation of Translated Small Open Reading Frames. Curr Protoc Chem Biol 11(4): e77.
- Ma, J., Diedrich, J. K., Jungreis, I., Donaldson, C., Vaughan, J., Kellis, M., Yates, J. R., 3rd and Saghatelian, A. (2016). Improved Identification and Analysis of Small Open Reading Frame Encoded Polypeptides. Anal Chem 88(7): 3967-3975.
- Magny, E. G., Platero, A. I., Bishop, S. A., Pueyo, J. I., Aguilar-Hidalgo, D. and Couso, J. P. (2021). Pegasus, a small extracellular peptide enhancing short-range diffusion of Wingless. Nat Commun 12(1): 5660.
- Orr, M. W., Mao, Y., Storz, G. and Qian, S. B. (2020). Alternative ORFs and small ORFs: shedding light on the dark proteome.Nucleic Acids Res 48(3): 1029-1042.
- Zhang, C., Zhou, B., Gu, F., Liu, H., Wu, H., Yao, F., Zheng, H., Fu, H., Chong, W., Cai, S., et al. (2022). Micropeptide PACMP inhibition elicits synthetic lethal effects by decreasing CtIP and poly(ADP-ribosyl)ation. Mol Cell 82(7): 1297-1312 e1298.
Article Information
Copyright
© 2023 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cao, X., Chen, Y., Khitun, A. and Slavoff, S. A. (2023). BONCAT-based Profiling of Nascent Small and Alternative Open Reading Frame-encoded Proteins. Bio-protocol 13(1): e4585. DOI: 10.21769/BioProtoc.4585.
Category
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









