- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Sclerotinia sclerotiorum Protoplast Preparation and Transformation
(*contributed equally to this work) Published: Vol 13, Iss 1, Jan 5, 2023 DOI: 10.21769/BioProtoc.4581 Views: 1611
Reviewed by: Zhibing LaiShweta PanchalRitu Gupta
Abstract
Sclerotinia sclerotiorum causes white mold, leading to substantial losses on a wide variety of hosts around the world. Many genes encoding effector proteins play important roles in the pathogenesis of S. sclerotiorum. Therefore, establishment of a transformation system for the exploration of gene function is necessarily significant. Here, we introduce a modified protocol to acquire protoplasts for transformation and generate knockout strains, which completements the transformation system of S. sclerotiorum.
Background
Sclerotinia sclerotiorum (Helotiales: Sclerotiniaceae), a phytopathogenic pathogen, infects more than 400 plants, including vegetables and ornamental plants (Kabbage et al., 2015). As the complete genome assembly of S. sclerotiorum is available, many genes encoding effector proteins have been preliminarily validated (Derbyshire et al., 2017). Preparation and transformation of protoplast are important measures to obtain mutants and research gene function. We introduce a protocol based on a standard protocol with minor modifications (Rollins, 2003). This method redounds to production of complete protoplasts for observation and transformation. Using this method, we successfully acquired SsVPS51, SsDCTN1, and SsSAC1 knockout mutants, respectively (Qiao et al., 2021). Therefore, polyethylene glycol–mediated transformation is suitable for protoplast transformation of S. sclerotiorum, providing a convenient method for studying its genetics.
Materials and Reagents
1.5 mL microfuge tube (Axygen)
2.0 mL microfuge tube (Axygen)
Petri dish (9 cm diameter)
40 µm cell strainer (Xiyan Co., Ltd., catalog number: 15-1040)
Syringe filter (Pall Corporation, catalog number: 4612-0.2 µm)
Sterile blade (ShangHaiLianHui Medical Instrument co., Ltd.)
50 mL centrifuge tube
50 mL conical flask
100 mL conical flask
250 mL conical flask
KCl (BIODEE, catalog number: DE-0395A-250 g)
CaCl2 (BIODEE, catalog number: DE-0556A-500 g)
Agar A (BBI Life Science, catalog number: A600010-500 g)
Potato dextrose broth (Solarbio Life Sciences, catalog number: P9240)
D-sorbitol (Sigma-Aldrich, catalog number: S3889-500 g)
Tris-HCl (Sigma-Aldrich, catalog number: T6666-500 g)
Sucrose (BBI Life Science, catalog number: A100335-250 g)
Yeast extract (BBI Life Science, catalog number: A515245-500 g)
Hygromycin B (Thermo Fisher Scientific, GibcoTM , catalog number: 10687010-20 mL)
Glucanex (Sigma-Aldrich, catalog number: L1412-5 g)
Polyethylene glycol 3500 (Sigma-Aldrich, catalog number: 25322-68-3-250 g)
Spermidine (Sigma-Aldrich, catalog number: 05292-1 mL)
Heparin (Sigma-Aldrich, catalog number: H3149-500 KU)
Casamino acid (Sangon Biotech, catalog number: A603060-100 g)
KCl buffer (pH = 6–6.5) (see Recipes)
0.5% Glucanex (see Recipes)
Phanta Super-Fidelity DNA Polymerase (Vazyme, catalog number: P501-d2)
STC solution (see Recipes)
SPTC solution (see Recipes)
PDA medium (see Recipes)
PDB medium (see Recipes)
RM medium (see Recipes)
RMA medium (see Recipes)
Equipment
1,000 µL micropipette (Eppendorf)
200 µL micropipette (Eppendorf)
10 µL micropipette (Eppendorf)
Centrifuge (Eppendorf, model: 5424R)
Centrifuge (Eppendorf, model: 5810R)
Hemocytometer (Devan Scientific Co., Ltd., catalog number: AP-0650010)
Bacterial shaker incubator (Crystal Technology & Industries, model: IS-AX-190L)
Biochemical incubator (Shanghai Yuejin Medical Device Co., Ltd, model: HPX-B400)
Zeiss Axio Observer (Zeiss)
Procedure
Culturing and enrichment of S. sclerotiorum mycelium
Cut the PDA medium (see Recipes) where S. sclerotiorum grew on for three days into mycelium blocks (about 1 mm length × 1 mm width) (Figure 1).
Note: The ideal cutting area is preferably at the edge of the fungal colony, where S. sclerotiorum hyphae actively grow.
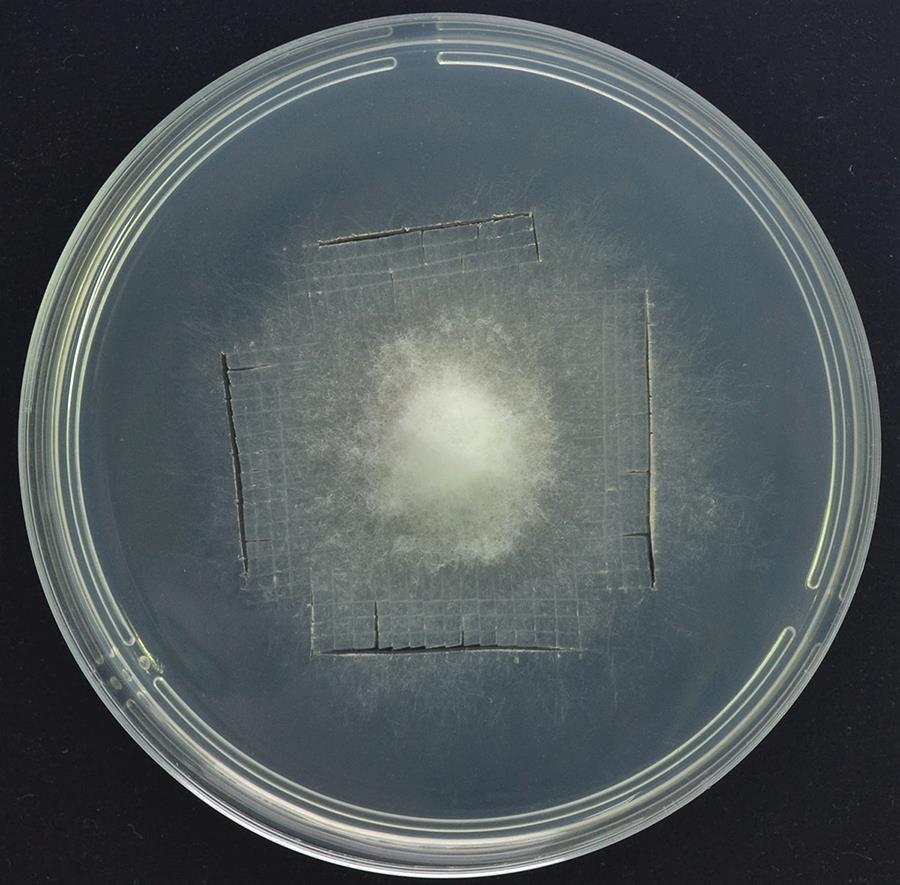
Figure 1. The ideal cutting areaCollect the mycelium blocks into a 250 mL conical flask containing 100 mL of PDB medium, and shake the flask to mix them together.
Note: It’s better to add approximately 200 mycelium blocks to 100 mL of PDB medium. This proportion guarantees that S. sclerotiorum uptakes adequate nutrition and produces enough fresh mycelium.
Fasten the conical flask on a shaking table (180 × g, 25 °C) and culture for 14–20 h, until fresh mycelium grows from the mycelium blocks (Figure 2).
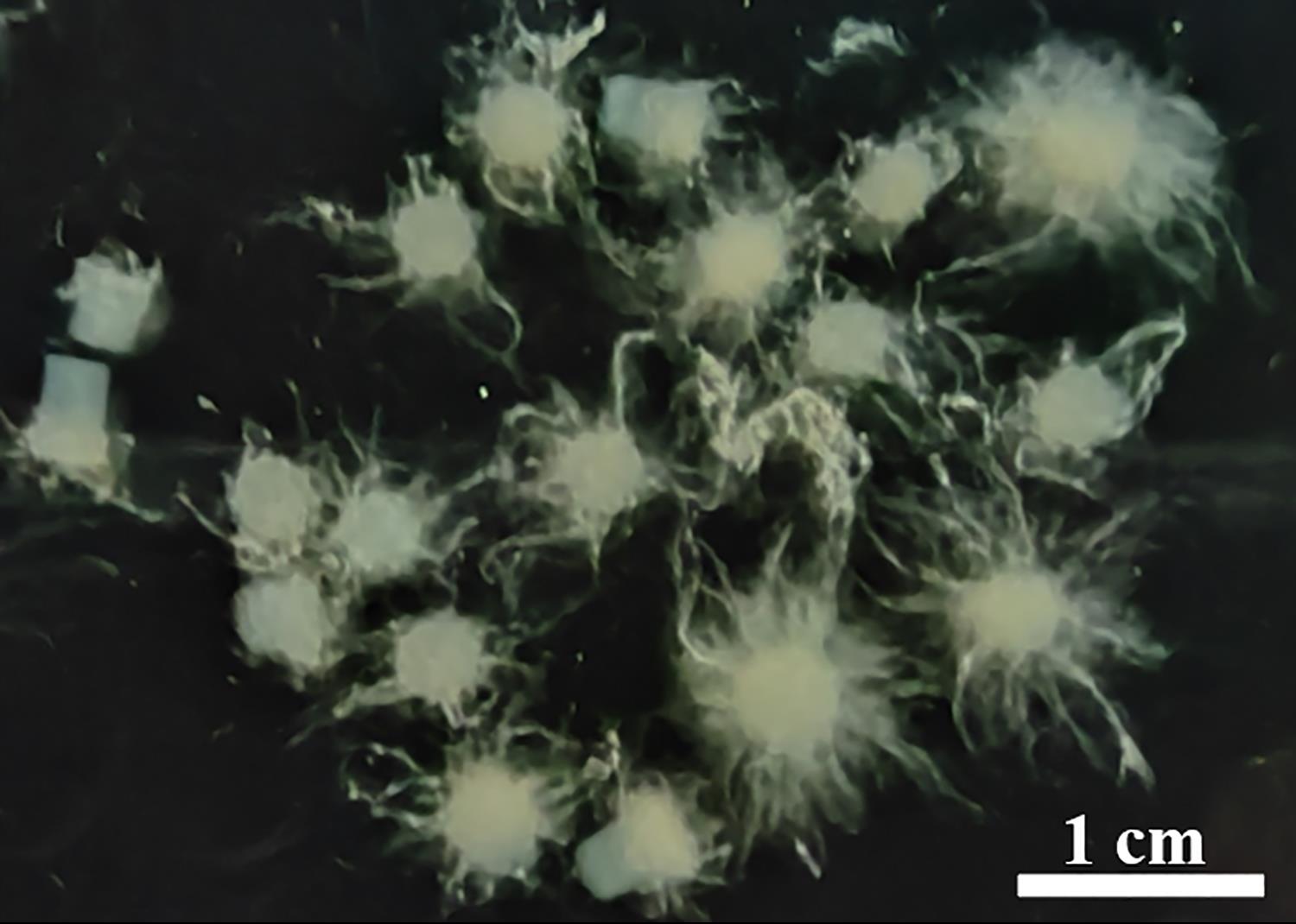
Figure 2. Fresh mycelium after culturing for 14–20 h. Scale bar = 1 cm.Using a tweezer, separate fresh mycelium from mycelium blocks, and collect them in a 50 mL centrifuge tube.
Resuspend the mycelium with 20 mL of KCl buffer, centrifuge at 1,500 × g and 25 °C for 5 min, and then discard the supernatant.
Repeat step A5 once again.
Protoplast preparation
Add 20 mL of 0.5% Glucanex [lysing enzyme from Trichoderma harzianum (Sigma)] to a 50 mL centrifuge tube containing 1 g of mycelium material. Incubate with gentle shaking (120 × g on the rotator) in the dark at room temperature for 2–3 h.
After the enzymatic digestion, filter the digestion mixture using a cell strainer, and collect the filtrate into a 50 mL centrifuge tube (Figure 3A).
Note: Place the filtrate on ice and proceed cautiously and gently with the next steps, as protoplasts are fragile and may easily rupture (Figure 3B).
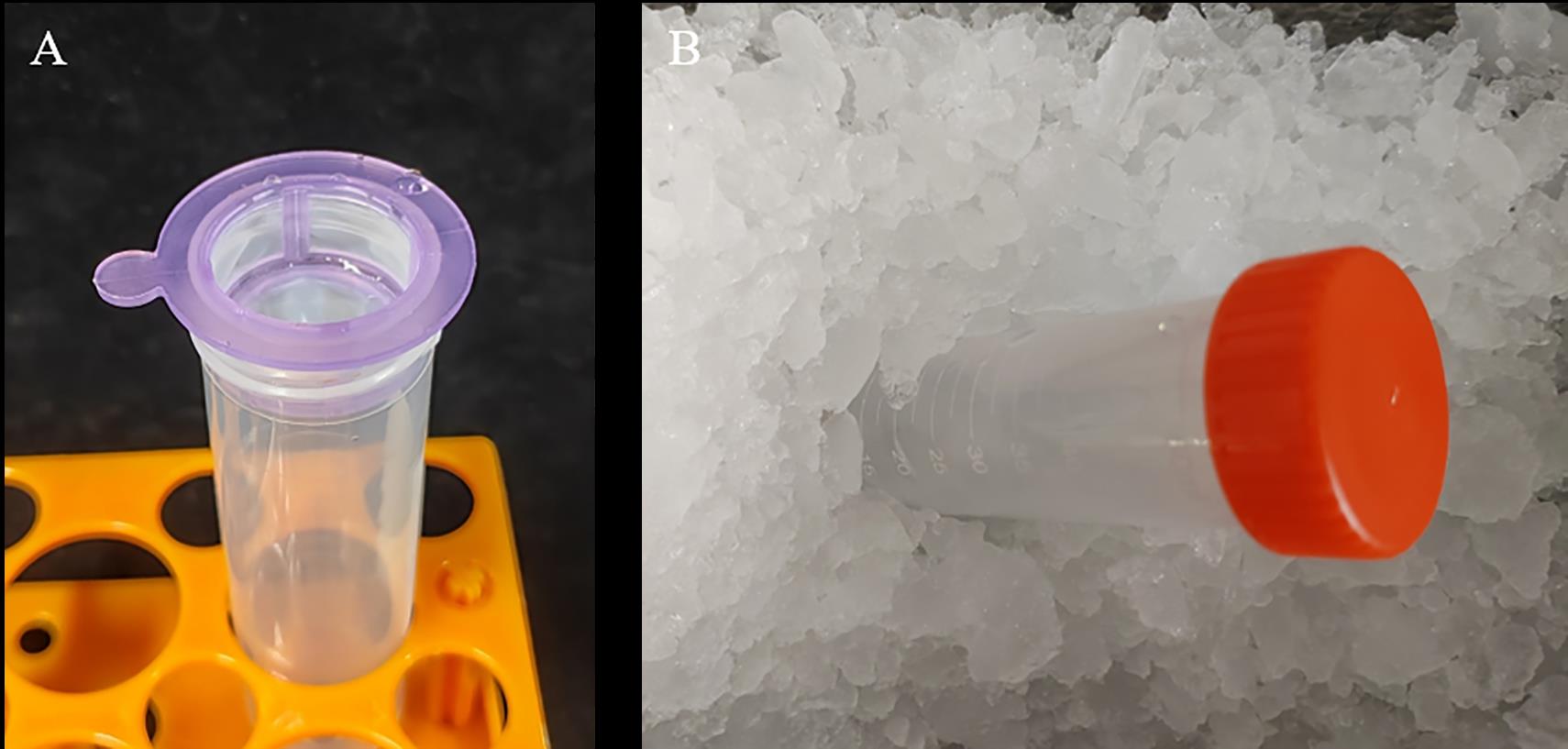
Figure 3. Filter the protoplasts and place the filtrate on iceCentrifuge the filtrate at 1,200 × g and 4 °C for 10 min, and carefully discard the supernatant.
Wash the protoplasts gently with 5 mL of precooled KCl buffer, centrifuge at 5,000 × g and 4 °C for 2 min, and then discard the supernatant carefully.
Repeat step B4 once again.
Note: Steps B4 and B5 effectively remove the residual enzyme mixture.
Resuspend the sediment with 1 mL of precooled STC, centrifuge at 5,000 × g and 4 °C for 2 min, and discard the supernatant by slowly decanting the tube.
Resuspend the sediment gently with 800 µL of precooled STC buffer and add 200 µL of precooled SPTC buffer drop by drop with a pipette.
Place the protoplast solution on ice and check the status of the protoplasts with a Zeiss Axio Observer (Figure 4).
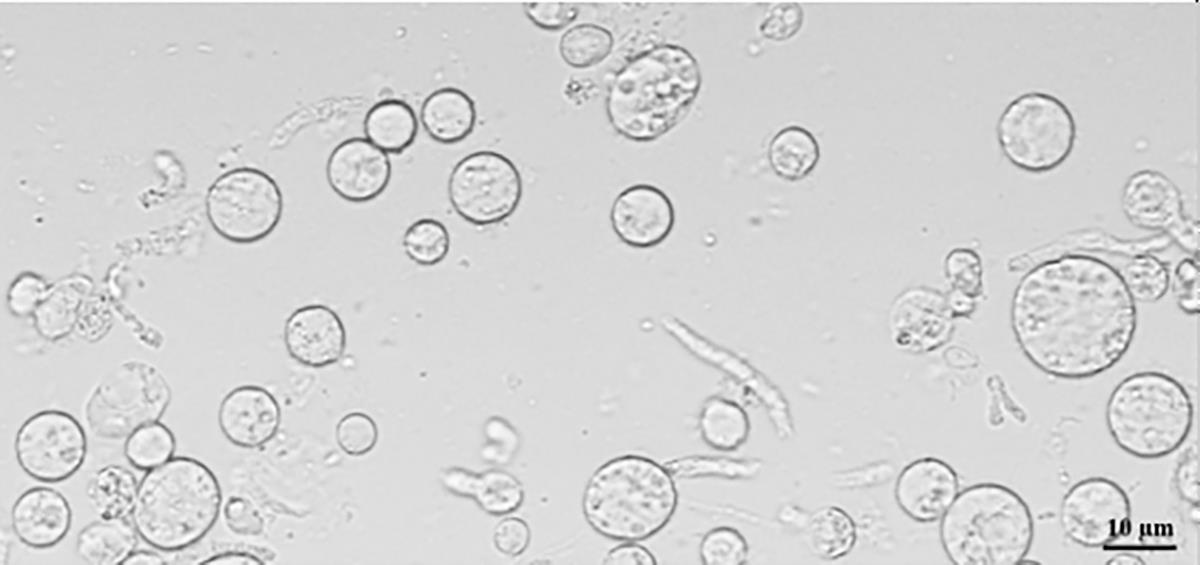
Figure 4. Protoplasts of S. sclerotiorum. Scale bar = 10 µm.Protoplast transformation
Resuspend the protoplasts to a concentration of 1 × 108 protoplast per milliliter, in four parts of STC and one part of SPTC.
Note: Calculate protoplast concentration with a hemocytometer, according to the manufacturer’s instructions.
Add 5 µg of transforming DNA, 5 µL of spermidine (50 mM stock), and 5 µL of heparin (5 mg/mL) to 100 µL of protoplast suspension in a 1.5 mL microfuge tube, and then mix gently.
Note: Mix them together by flicking the bottom of the microfuge tube instead of using a pipette.
Incubate the mixture on ice for 30 min, then gently add 1 mL of SPTC solution, and incubate at room temperature for 20 min.
Transfer the transformation mixture into a 50 mL centrifuge tube, and add 10–20 mL of Regeneration Medium (RM).
Culture the mixture with 100 × g agitation, at 25 °C overnight.
Gently mix the protoplasts with 200 mL of RMA medium at 43 °C, pour this into Petri dishes (10 mL per dish), and incubate at 25 °C for 24 h.
Note: Heat up RMA medium until it melts completely, and cool it down in a water bath set to 43 °C.
After, cover the plates with 10 mL of RMA medium containing 100 µg/mL hygromycin B.
Culture at 25 °C for 7–10 days, and transfer the transformants to fresh PDA medium containing 100 µg/mL hygromycin B.
Transfer the hyphal tip of hygromycin-resistant transformants to PDA medium containing 100 µg/mL hygromycin B. Repeat the selection at least three times (Figure 5A).
Note: Pick the edge hyphae of recovered hygromycin-resistant transformants using a sterile blade for the first selection, and cut the mycelium block for the next two selections.
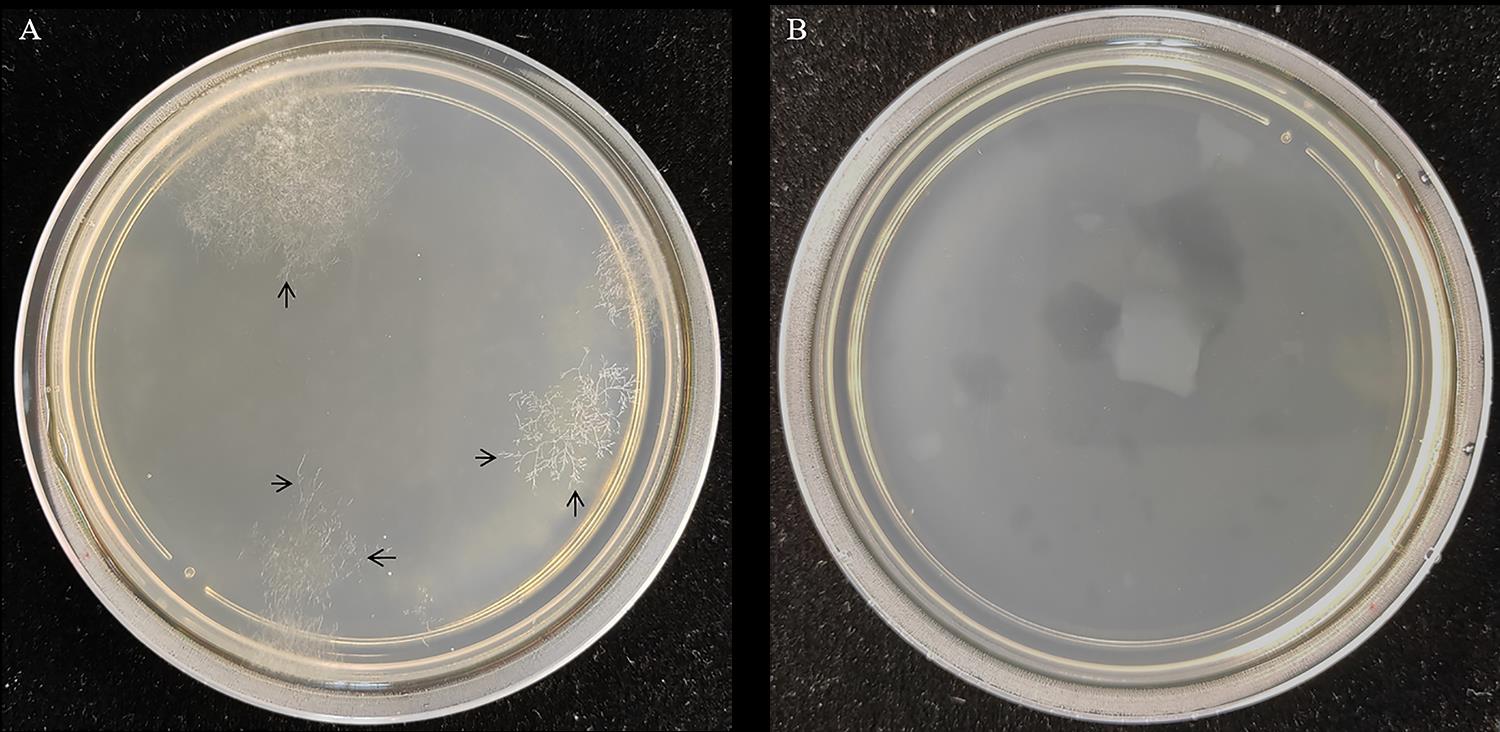
Figure 5. Colony morphology of S. sclerotiorum grown on selective RMA medium. A. Colony morphology denoting successful transformation. The hyphal tips labeled by black arrows can be transferred to PDA medium for selection. B. Negative control plate made by adding no DNA in step C2.Extract total RNA of any colony and reverse transcript into cDNA. Perform PCR to validate positive colony by Phanta Super-Fidelity DNA Polymerase with the following cycles:
Pre-denature the cDNA at 95 °C for 30 s
Denature the cDNA at 95 °C for 30 s
50 °C for 15 s
72 °C for 90 s
Repeat steps b-d for 25–35 cycles
72 °C for 10 min
Note: Primers designed to check mutants are as follows:
SsVPS51-GT-5’-F: CTTAGAGACTTGAGGAAGTC
SsVPS51-GT-3’-R: CTACTTCTATTGCTATTC
Recipes
KCl buffer (pH =6–6.5, sterilize by syringe filter, and store at 25 °C)
Reagent Final concentration Amount KCl 0.6 M 4.47 g CaCl2 50 mM 0.56 g ddH2O n/a up to 100 mL Total n/a 100 mL 0.5% Glucanex (store at 4 °C)
Reagent Final concentration Amount Glucanex n/a 0.5 g KCl buffer n/a up to 100 mL Total n/a 100 mL STC solution (autoclave at 121 °C for 20 min)
Reagent Final concentration Amount Sorbitol 0.8 M 145.74 g Tris-HCl (pH = 8.0) 50 mM 7.88 g CaCl2 50 mM 5.54 g ddH2O n/a up to 1,000 mL Total n/a 1,000 mL SPTC solution (autoclave at 121 °C for 20 min)
Reagent Final concentration Amount Sorbitol 0.8 M 145.74 g Tris-HCl (pH = 8.0) 50 mM 7.88 g CaCl2 50 mM 5.54 g Polyethylene glycol 3500 0.11 M 400 g ddH2O n/a up to 1,000 mL Total n/a 1,000 mL PDA medium (autoclave at 121 °C for 20 min)
Reagent Final concentration Amount Potato dextrose broth n/a 24 g Agar A n/a 15 g ddH2O n/a up to 1,000 mL Total n/a 1,000 mL PDB medium (autoclave at 121 °C for 20 min)
Reagent Final concentration Amount Potato dextrose broth n/a 24 g ddH2O n/a up to 1,000 mL Total n/a 1,000 mL RM medium (autoclave at 121 °C for 20 min)
Reagent Final concentration Amount Sucrose n/a 274 g Yeast extract n/a 1 g Casamino acid n/a 1 g ddH2O n/a up to 1,000 mL Total n/a 1,000 mL RMA medium (autoclave at 121 °C for 20 min)
Reagent Final concentration Amount Sucrose n/a 274 g Yeast extract n/a 1 g Casamino acid n/a 1 g Agar A n/a 15 g ddH2O n/a up to 1,000 mL Total n/a 1,000 mL
Acknowledgments
This work was supported by the National Natural Science Foundation of China (32072404) and the Natural Science Foundation of Jiangsu Province (BK20211524). We thank Jeffrey A. Rollins for his work in establishing this protocol.
Competing interests
No financial, personal, or professional interests have influenced the work.
References
- Derbyshire, M., Denton-Giles, M., Hegedus, D., Seifbarghy, S., Rollins, J., van Kan, J., Seidl, M. F., Faino, L., Mbengue, M., Navaud, O., et al. (2017). The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol Evol 9(3): 593-618.
- Kabbage, M., Yarden, O. and Dickman, M. B. (2015). Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic lifestyle.Plant Sci 233: 53-60.
- Qiao, L., Lan, C., Capriotti, L., Ah-Fong, A., Nino Sanchez, J., Hamby, R., Heller, J., Zhao, H., Glass, N. L., Judelson, H. S., et al. (2021). Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol Journal 19(9): 1756-1768.
- Rollins, J. A. (2003). The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Molecular Plant-Microbe Interactions 16: 785-795.
Article Information
Copyright
© 2023 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lan, C., Qiao, L. and Niu, D. (2023). Sclerotinia sclerotiorum Protoplast Preparation and Transformation. Bio-protocol 13(1): e4581. DOI: 10.21769/BioProtoc.4581.
Category
Microbiology > Microbial cell biology
Molecular Biology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









