- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Direct in vitro Fatty Acylation Assay for Hedgehog Acyltransferase
Published: Vol 12, Iss 24, Dec 20, 2022 DOI: 10.21769/BioProtoc.4573 Views: 1316
Reviewed by: Gal HaimovichQin TangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Purification of Native Acetyl CoA Carboxylase From Mammalian Cells
Yaxue Sun [...] Hongtao Zhu
Feb 20, 2025 2168 Views
Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2256 Views
Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 385 Views
Abstract
Several assays have been developed to monitor the in vitro catalytic activity of Hedgehog acyltransferase (Hhat), an enzyme critical to the Hedgehog signaling pathway in cells. However, the majority of these previously reported assays involve radioactive fatty acyl donor substrates, multiple steps to achieve product readout, or specialized equipment. To increase safety, efficiency, and convenience, we developed a direct, fluorescent in vitro assay to monitor Hhat activity. Our assay utilizes purified Hhat, a fluorescently labeled fatty acyl-CoA donor substrate, and a Sonic hedgehog (Shh) peptide recipient substrate sufficient for fatty acylation. The protocol is a straightforward process that yields direct readout of fatty acylated Shh peptide via fluorescence detection of the transferred fatty acyl group.
Graphical abstract

Graphical abstract adapted from Schonbrun and Resh (2022)
Background
The Hedgehog (Hh) pathway plays an essential role in embryonic development and in multiple cancers in adults. Signaling is mediated by the secretion of Hh proteins, e.g., Sonic hedgehog (Shh), ultimately resulting in the activation of GLI transcription factors and expression of Hh pathway target genes. An essential component of the signaling pathway is the enzyme Hedgehog acyltransferase (Hhat), which catalyzes the transfer of palmitate as well as other fatty acyl groups to the N-terminal cysteine of Hh proteins (Schonbrun and Resh, 2022). This post-translational modification has been shown to be required for Hh signaling (Micchelli et al., 2002;Chen et al., 2004).
Hhat is a member of the membrane-bound O-acyltransferase enzyme family and is localized in the membrane of the endoplasmic reticulum. To date, several assays have been reported to measure the catalytic activity of Hhat in vitro. However, some of these assays rely on the use of radioactive substrates (Buglino and Resh, 2008; Jiang et al., 2021), which have safety hazards and require specialized waste disposal. Others require specialized equipment (Lanyon-Hogg et al., 2017) or multiple steps (Lanyon-Hogg et al., 2015) to achieve product readout, or measure a change in fluorescence anisotropy conferred by fatty acylation of a fluorescently conjugated Shh peptide (Lanyon-Hogg et al., 2019; Andrei et al., 2022). We set out to develop a straightforward, fluorescence-based assay to quantify the catalytic activity of purified Hhat in vitro by directly monitoring the transfer of a fluorescently labeled fatty acid to Shh. This protocol has the advantage of allowing for unambiguous competition analysis with other fatty acyl-CoAs (in addition to competition analysis with alternative forms of Shh substrate), by including these unlabeled substrates in the reaction. The two labeled substrates, biotinylated Shh peptide and NBD-labeled fatty acyl-CoAs (as well as the corresponding, unlabeled compounds for competition analysis) are readily obtained from commercial sources. Our assay is sensitive and specific and can potentially be adapted to a high-throughput format using a streptavidin-coated multi-well plate to capture the biotinylated Shh substrate.
Materials and Reagents
0.5 mL tubes (RPI, catalog number: 145505)
Black side, transparent bottom 96-well plate (Thermo Scientific, catalog number: 265301)
Biotinylated Shh peptide [CGPGRGFGKR-(PEG2)-K(Biotin)-NH2] [Anaspec and Peptide 2.0, custom synthesis, PEG2 = Fmoc-NH-PEG2-CH2-COOH; M.W. (theoretical) 1532.82 g/mol, order at least 5 mg, free N-terminus, NH2 at C-terminus, trifluoroacetate salt, QC analysis by MS and >95% purity by peak area on HPLC (220 nm, C18 column, Buffer A: 0.05% TFA in H2O, Buffer B: 0.05% TFA in 90% CH3CN, linear gradient 10%–35% B in 25 min] (store at a minimum of -20 °C)
Nitrobenzoxadiazole (NBD)-fatty acyl-CoAs (Avanti, catalog numbers: 810229P and 810705P) (store aliquots at a minimum of -20 °C)
n-Dodecyl-β-D-maltopyranoside (DDM) (Anatrace, catalog number: D310) (store at -20 °C)
3× FLAG® peptide (Millipore Sigma, catalog number: F4799) (store at -20 °C)
n-Octyl-β-D-glucopyranoside (OG) (EMD Millipore, catalog number: 494459) (store at room temperature)
Triton X-100 (Fisher Scientific, catalog number: BP151-500) (store at room temperature)
MES (Fisher Scientific, catalog number: BP300-100) (store at room temperature)
DTT (Promega, catalog number: V3151) (store aliquots at -20 °C)
PierceTM high-capacity streptavidin beads (Thermo Scientific, catalog number: 20361) (store at 4 °C)
Purified Hhat and mock purified eluate or elution buffer (see Recipes) (Schonbrun and Resh, 2022) (store at -80 °C)
Reaction buffer (see Recipes)
Wash buffer (see Recipes)
Equipment
Fluorescence plate reader (BioTek Synergy H1)
Refrigerated tabletop microcentrifuge (Eppendorf Centrifuge, model: 5417R)
PicoFuge (Stratagene)
Vacuum aspirator with attached 26½ G needle (Becton Dickinson, catalog number: 305111)
Repeat or regular pipettors
Software
Gen5 (BioTek)
Excel (Microsoft)
Prism (GraphPad)
Procedure
Assay setup
Set up the appropriate number of 0.5 mL plastic tubes in a rack at room temperature. Reactions should be performed in at least duplicate.
Prepare fresh reaction buffer (room temperature) by adding DTT prior to the start of the assay.
Prepare a suspension of streptavidin-coated beads in wash buffer.
Transfer a total volume of slurry corresponding to 15 µL of slurry per reaction into a tube. Example: In an assay with eight reactions to be assayed in duplicate, 240 µL [(8 × 2) × 15 µL] of slurry should be prepared along with an additional 60 µL (4 × 15 µL) for the standards, to make a total of 300 µL slurry (240 µL + 60 µL). The combined slurry should be resuspended with 1,000 µL [(16 + 4) × 50 µL] wash buffer after washing. Be sure to include in the calculation for the beads the additional beads needed for NBD standards at the end of the assay (see step C5).
Wash the beads once by adding a total volume of wash buffer corresponding to 50 µL per reaction to the tube and inverting the tube several times.
Centrifuge the beads at 2,700 × g and 4 °C for 1 min in a tabletop microcentrifuge and aspirate the supernatant using a 26½ G needle.
Resuspend the beads with a total volume of wash buffer corresponding to 50 µL of wash buffer per reaction.
Keep tube on ice.
Aliquot reaction buffer and the desired concentrations of NBD-fatty acyl-CoA (we typically use 10 μM) and Shh peptide (we typically use 10 μM) into each tube. For inhibition or substrate competition assays, include drug/inhibitor [e.g., RU-SKI 201 (Cayman, catalog number: 21101)] or unlabeled substrates (e.g., non-fluorescent fatty acyl-CoA or non-biotinylated Shh protein/peptide) at desired concentrations. Include appropriate control reactions.
Note: The final volume in each tube should be 100 µL, and importantly, should include a final concentration of 0.1% DDM (detergent component of the purified Hhat eluate). Final concentrations of MES, OG (w/v), and DTT from the reaction buffer should be in the range of 125 mM, 0.06%, and 750 µM, respectively. If desired, a master mix of reaction buffer and substrates can be prepared.
Reaction
Initiate the reaction by adding purified Hhat enzyme (or equivalent volume of mock purified eluate or elution buffer as a negative control) (we typically use 130 nM Hhat). Briefly vortex and pulse-spin in a PicoFuge to collect the sample at the bottom of the tube.
Immediately incubate the reactions in the dark at 37 °C for the desired amount of time.
Note: For kinetic studies, a time course must be performed first to determine the linear range of the reaction. Suggested time points for initial kinetic studies are 0, 10, 20, 40, 60, and 90 min.
Quench the reactions by placing the tubes on ice and adding 60 µL of ice-cold bead suspension.
Isolation and quantification of reaction products
Incubate tubes in the dark with rocking at 4 °C for 1 h.
Centrifuge the beads at 2,700 × g and 4 °C for 1 min in a tabletop microcentrifuge and carefully aspirate the supernatant using a 26½ G needle.
Wash the beads three times with 100 µL of wash buffer. Between each wash, spin tubes and aspirate supernatant as above.
Tip for washing: place the tubes in a single rack, cover the top of the rack, and invert the rack approximately five times to wash.
After the final wash, carefully aspirate the supernatant, resuspend the beads in each tube with 60 µL of wash buffer, and carefully transfer the beads from each tube to individual wells of a black side, transparent bottom 96-well plate. Use a fresh pipette tip for each tube.
Note: To maximize bead transfer, after the first transfer of beads into the plate, use the pipette tip to push residual beads from the sides down to the bottom of the tube where some suspension remains and transfer this remaining suspension into the same well.
To obtain a conversion factor of NBD fluorescence (RFU) to pmol of NBD-labeled product for data quantification (see next section), load standards [known amounts (e.g., 500–2,000 pmol)] of NBD-acyl-CoA in bead suspension (60 µL, as in step C4) into separate wells of the plate.
Read NBD fluorescence (RFU) in a plate reader at excitation/emission wavelengths of 465/535 nm.
Data analysis
Export RFU readings from Gen5 software directly to an Excel spreadsheet.
Quantify NBD-acylated Shh product using the RFU-to-pmol conversion factor obtained from the standards. To obtain the conversion factor, divide the RFU output value by the known number of pmol of NBD donor loaded. For calculation of the initial rate, choose a time point at which the reaction is linear (we typically use 10 min). Divide pmol of NBD-acylated Shh produced at that time point by the number of minutes, then divide by the number of pmol of Hhat to give pmol NBD-acylated Shh per min per pmol Hhat in units of min-1 .
Data can be plotted in Excel or GraphPad Prism with a standard bar or line graph, or in GraphPad Prism with best-fit kinetic models, as appropriate (e.g., Figure 1).
Note: For kinetic models, data should be presented as an initial rate (pmol product per min or pmol product per min per amount of enzyme).
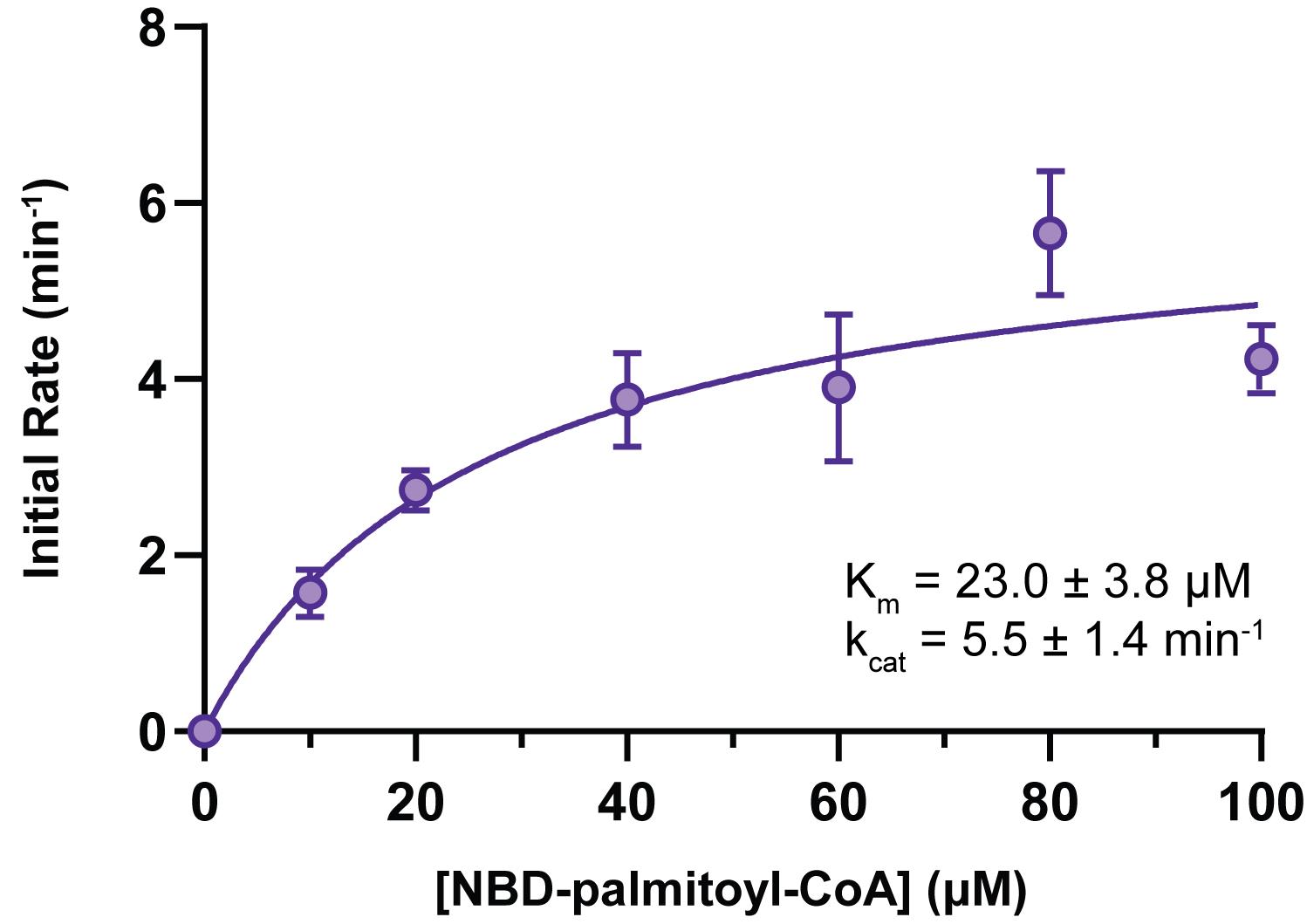
Figure 1. Hhat enzymatic activity as a function of NBD-palmitoyl-CoA concentration. Increasing concentrations of NBD-palmitoyl-CoA were incubated with 25 µM wild-type Shh peptide and purified Hhat for 10 min at 37°C. The initial rate of Hhat-mediated NBD-palmitoylation of Shh was quantified; Michaelis–Menten fit of the data (produced in GraphPad Prism) is shown with apparent kinetic parameters as insets (Schonbrun and Resh, 2022).
Recipes
Elution buffer
20 mM HEPES, pH 7.5
350 mM NaCl
1% (v/v) glycerol
1% (w/v) DDM
150 µg/mL 3× FLAG® peptide
Reaction buffer
167 mM MES, pH 6.5
0.083% (w/v) OG
1 mM DTT (added fresh)
Wash buffer
167 mM MES, pH 6.5
0.083% (v/v) Triton X-100
Acknowledgments
This research was supported by NIH Grant GM116860, by a grant from the Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and The Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center, by Cancer Center Core Support Grant P30 CA008748 from the National Institutes of Health to Memorial Sloan Kettering Cancer Center, and by an American Heart Association Predoctoral Fellowship #20PRE35040000 (2020) to ARS.
Competing interests
The authors declare they have no competing interests with the contents of this article.
References
- Andrei, S. A., Tate, E. W. and Lanyon-Hogg, T. (2022). Evaluating Hedgehog Acyltransferase Activity and Inhibition Using the Acylation-coupled Lipophilic Induction of Polarization (Acyl-cLIP) Assay. Methods Mol Biol 2374: 13-26.
- Buglino, J. A. and Resh, M. D. (2008). Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J Biol Chem 283(32): 22076-22088.
- Chen, M. H., Li, Y. J., Kawakami, T., Xu, S. M. and Chuang, P. T. (2004). Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev 18(6): 641-659.
- Jiang, Y., Benz, T. L. and Long, S. B. (2021). Substrate and product complexes reveal mechanisms of Hedgehog acylation by HHAT. Science 372(6547): 1215-1219.
- Lanyon-Hogg, T., Masumoto, N., Bodakh, G., Konitsiotis, A. D., Thinon, E., Rodgers, U. R., Owens, R. J., Magee, A. I. and Tate, E. W. (2015). Click chemistry armed enzyme-linked immunosorbent assay to measure palmitoylation by hedgehog acyltransferase. Anal Biochem 490: 66-72.
- Lanyon-Hogg, T., Patel, N. V., Ritzefeld, M., Boxall, K. J., Burke, R., Blagg, J., Magee, A. I. and Tate, E. W. (2017). Microfluidic Mobility Shift Assay for Real-Time Analysis of Peptide N-Palmitoylation. SLAS Discov 22(4): 418-424.
- Lanyon-Hogg, T., Ritzefeld, M., Sefer, L., Bickel, J. K., Rudolf, A. F., Panyain, N., Bineva-Todd, G., Ocasio, C. A., O'Reilly, N., Siebold, C., Magee, A. I. and Tate, E. W. (2019). Acylation-coupled lipophilic induction of polarisation (Acyl-cLIP): a universal assay for lipid transferase and hydrolase enzymes. Chem Sci 10(39): 8995-9000.
- Micchelli, C. A., The, I., Selva, E., Mogila, V. and Perrimon, N. (2002). Rasp, a putative transmembrane acyltransferase, is required for Hedgehog signaling. Development 129(4): 843-851.
- Schonbrun, A. R. and Resh, M. D. (2022). Hedgehog acyltransferase catalyzes a random sequential reaction and utilizes multiple fatty acyl-CoA substrates. J Biol Chem 289(10): 102422.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Schonbrun, A. R. and Resh, M. D. (2022). A Direct in vitro Fatty Acylation Assay for Hedgehog Acyltransferase. Bio-protocol 12(24): e4573. DOI: 10.21769/BioProtoc.4573.
Category
Cancer Biology > Cancer biochemistry > Protein
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









