- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of Alternative End-Joining in HNSC Cell Lines Using DNA Double-Strand Break Reporter Assays
Published: Vol 12, Iss 17, Sep 5, 2022 DOI: 10.21769/BioProtoc.4506 Views: 2353
Reviewed by: Pilar Villacampa AlcubierreHongLok LungAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Conditional Human BRD4 Knock-In Transgenic Mouse Genotyping and Protein Isoform Detection
Michael Paul Lewis [...] Cheng-Ming Chiang
Apr 5, 2022 3594 Views
OxiDIP-Seq for Genome-wide Mapping of Damaged DNA Containing 8-Oxo-2'-Deoxyguanosine
Francesca Gorini [...] Stefano Amente
Nov 5, 2022 2859 Views
Conditional Depletion of STN1 in Mouse Embryonic Fibroblasts
Sara Knowles and Weihang Chai
Apr 20, 2024 4312 Views
Abstract
The main cellular pathways to repair DNA double-strand breaks (DSBs) and protect the integrity of the genome are homologous recombination (HR), non-homologous end-joining (NHEJ), and alternative end-joining (Alt-EJ). Polymerase theta-regulated Alt-EJ is an error-prone DSB repair pathway characterized by microhomology usage. Considering its importance in cancer treatment, technologies for detection of Alt-EJ in cancer cells may facilitate the study of the mechanisms of carcinogenesis and the development of new therapeutic targets. DSB reporter assay is the classical method for detecting Alt-EJ, which is primarily based on components of EJ2-puro cassette integration, I-SceI cleaving, and flow cytometry analysis. Here, we described an assay based on a modified I-Scel plasmid that can screen head and neck squamous cell carcinoma (HNSC) cells that were successfully transfected using selection medium with hygrovetine. We expect that this protocol will improve the fidelity and accuracy of reporter assays.
Graphical abstract:

Schematic overview of the workflow for establishment of Alt-EJ reporters.
Background
DNA double-strand breaks (DSBs) are among the most dangerous forms of DNA lesions and must be efficiently and accurately repaired to prevent genomic instability, tumorigenesis, or cell death (Rahimian et al., 2020; Huang et al., 2021). To deal with endogenous and exogenous threats, cells have evolved multiple DSB repair pathways, mainly including homologous recombination (HR), non-homologous end-joining (NHEJ), and alternative end-joining (Alt-EJ) (Mateos-Gomez et al., 2015; Liu et al., 2019; Wang et al., 2021; Fujii et al., 2022). Of these, Alt-EJ is an error-prone form of DSB repair that leads to chromosomal rearrangements and genomic instability (Liu et al., 2019; Caracciolo et al., 2021; Valikhani et al., 2021). It has been reported that Alt-EJ plays a crucial role in BRCA2-deficient cancers, human papillomavirus-associated cancers, and tumors deficient in the TGF-β signaling pathway (Ceccaldi et al., 2015; Liu et al., 2018, 2019, 2021, 2022; Guix et al., 2022). However, there are still many unknown aspects of Alt-EJ, such as the mechanisms for Alt-EJ regulation, Alt-EJ in tumor development, and DNA damage responses. To further explore the relevant mechanisms and target Alt-EJ for therapeutic benefits, the methods for Alt-EJ detection need to be established and standardized.
Reporter assays, proposed by Gunn et al. (2012), are the most commonly used tools for measuring Alt-EJ (Gunn et al., 2012). EJ2GFP-puro, a non-functional fluorescent protein expression cassette, needs to be integrated into the genome of the cells to be tested. Following the generation of a DSB in the cassette by I-SceI, the expression of the green fluorescent protein is resumed if the cell adopts Alt-EJ for repairing this DSB (Bennardo et al., 2008) (Figure 1). However, due to a lack of suitable screening conditions, the results are inevitably influenced by the efficiency of transfection. Here, we utilized another retroviral system with antibiotic resistance to make DSB and screen for target cells, a method adapted from Muraki et al. (2013). The results are presented in our recent publications (Liu et al., 2018, 2021). This method can avoid the influence of transfection efficiency on the experimental results and improve the accuracy and reproducibility of Alt-EJ study.
Our protocol has been successfully applied in the HNSC cell line SAS, as well as in the glioblastoma cell line U251 (Liu et al., 2018). Theoretically, this protocol could also be applied in other tumor or normal cell lines that require detection of Alt-EJ levels, but further validation is still needed. We can judge the mutational potential of cell chromosomes by testing the levels of Alt-EJ in sorted cells, and thus further explore the development and treatment of relevant cancers.
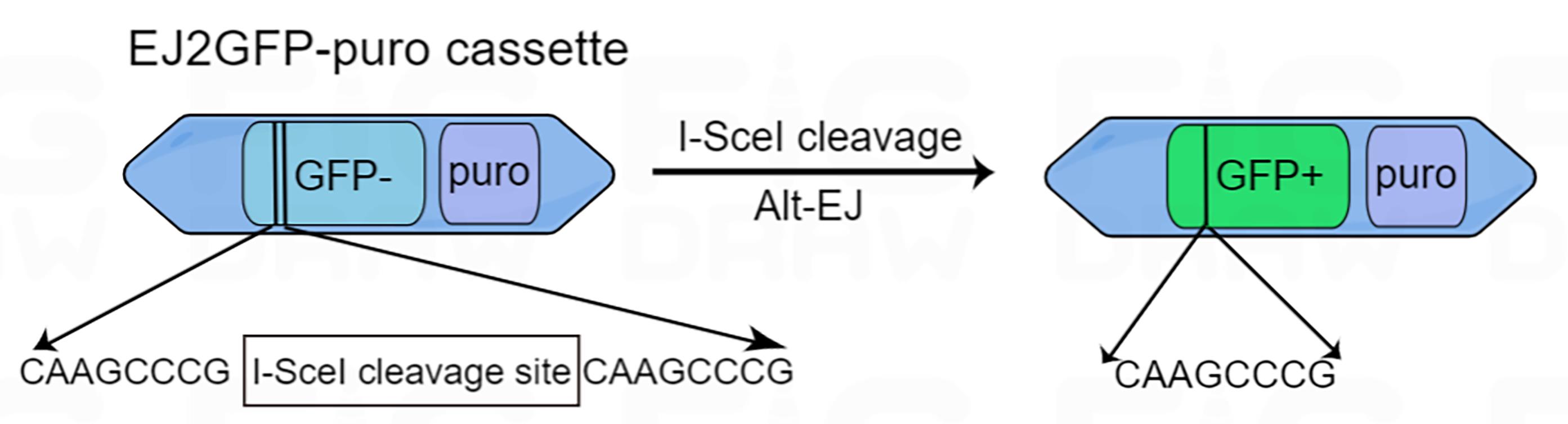
Figure 1. Diagram of the EJ2GFP-puro cassette. EJ2-GFP contains a GFP expression cassette that is disrupted by the I-SceI cleavage site. Repair by Alt-EJ results in restoration of GFP protein expression.
Materials and Reagents
Pipette tips 10 μL, 200 μL, and 1,250 μL (BEAVER, catalog numbers: 43140, 43143, 43152)
Pipettes set (Gilson, catalog numbers: GFAM00140)
Centrifuge tubes 15 mL and 50 mL (BEAVER, catalog numbers: 43008, 43009)
EP tube 1.5 mL (AXYGEN, catalog numbers: MCT-150-C-S)
Cell culture plate 6-well, 48-well, 24-well, and 96-well (CORNING, catalog numbers: 3516, 3548, 3526, 3599)
100 mm cell culture plates (BEAVER, catalog numbers: 43701)
HNSC cell line SAS (RIKEN BRC, catalog numbers: RCB1974)
Human embryonic kidney cell line HEK293T (ATCC, catalog number: CRL-3216)
Bovine growth serum (HyClone, catalog number: SH30541.03)
Dulbecco’s modified Eagle’s medium, DMEM (Gibco, catalog number: 31966047)
Streptomycin-penicillin (Gibco, catalog number: 15140122)
Trypsin-EDTA (Gibco, catalog number: 25300096)
Lipofectamine 2000 (Invitrogen, catalog number: 11668019)
Opti-MEM (Gibco, catalog number: 51985026)
EJ2GFP-puro (Addgene, catalog number: 44025)
pQCXIH-I-SceI (a gift from Dr. John P. Murnane, University of California San Francisco)
Puromycin (Sigma-Aldrich, catalog number: P8833)
Hygrovetine (Selleck, catalog number: S2908)
Round-bottom tubes with cell strainer cap, 5 mL (Falcon, catalog number: 38030)
LY2157299 (SelleckChem, catalog number: S2230)
Olaparib (LC Laboratories, catalog number: O-9201)
PBS (see Recipes)
SAS/HEK 293T medium (see Recipes)
Equipment
Cell culture incubator (Thermo Fisher Scientific, catalog number: 51026283)
Neubauer counting chamber (Carl Roth, catalog number: PC72.1)
Centrifuge (Eppendorf, catalog numbers: 5804)
CytoFLEX flow cytometer (Beckman Coulter, catalog numbers: B49008AC)
Software
FlowJo software for FACS analysis
Procedure
Stable integration of EJ2GFP-puro into SAS cells
Twenty-four hours prior to transfection, plate SAS cells in a 6-well plate at 5 × 105 cells per well. Each well contains 2 mL of SAS medium.
For each well, prepare one Mix A and one Mix B, each in a 1.5 mL tube. Mix gently with finger flicking and incubate for 5 min.
Component Mix A Mix B EJ2GFP-puro 2 μg - Lipofectamine 2000 - 5 μL Opti-MEM To 125 μL 120 μL Total 125 μL 125 μL Add Mix A to Mix B, mix gently, and incubate at room temperature for at least 20 min.
After washing the cells twice with 2 mL of 1× PBS, aspirate the PBS.
After adding the mixture to SAS cells, incubate at 37 °C and 5% CO2.
After 6 h of incubation, replace the mixture with SAS medium containing 2 mg/mL of puromycin.
Change the SAS medium containing 2 mg/mL of puromycin every 2–3 days until the cells reach 80%–90% confluence.
Trypsinize cells with trypsin-EDTA and count them.
Mix 60 cells with fresh SAS medium (containing 2 mg/mL of puromycin) to form a 6 mL cell suspension.
Add 100 μL of cell suspension to each well of a 96-well plate (100 μL of PBS for wells at the edge).
Four hours after plating, record the well with only one cell for subsequent screening.
When confluence exceeds 50% of well area, seed cells progressively into 48-well plates, 24-well plates, and 6-well plates. Select the cell line in the best condition for amplification and subsequent experiments. As there is a possibility that the selected monoclonal cell line is puromycin resistant and not EJ2GFP-puro successfully transfected, other monoclonal EJ2-puro-SAS cells will also need to be frozen for reserve.
Generation of I-SceI-induced DSBs
Retrovirus preparation
One day before packaging, plate 5 × 106 HEK 293T cells on a 100 mm cell culture plate containing 10 mL of HEK293T medium.
Prepare one Solution A and one Solution B, each in a 1.5 mL tube. Mix gently and incubate for 5 min.
Component Solution A Solution B pQCXIH-I-SceI 20 μg - Lipofectamine 2000 - 75 μL Opti-MEM To 1,250 μL 1,125 μL Total 1,250 μL 1,250 μL Mix Solution A and Solution B together and let it sit at room temperature for at least 20 min.
Wash the HEK 293T twice with 10 mL of 1× PBS.
Add the mixture (Solution A and Solution B) to HEK 293T and incubate for 6 h at 37 °C and 5% CO2.
After 6 h of incubation, remove the transfection medium and replace with fresh HEK293T medium. Continue to incubate at 37 °C and 5% CO2.
After 72 h, harvest virus-containing medium. Virus-containing medium can be stored temporarily at 4 °C for 3 days or in a -80 °C refrigerator for approximately 6 months.
Generation of DSBs using I-SceI in SAS cells
One day before infection, plate monoclonal EJ2-puro-SAS in a 6-well plate (containing 2 mL of SAS medium) at 5 × 105 cells per well.
Incubate the monoclonal reporter cells with a mixture of 1 mL of virus-containing medium and 1 mL of SAS medium for 24 h.
After 24 h, select infected cells with fresh SAS medium containing 400 mg/mL hygromycin for 10 days. Refresh the medium every 2–3 days. GFP-expressing cells are clearly visible under fluorescent microscopy (Figure 2).
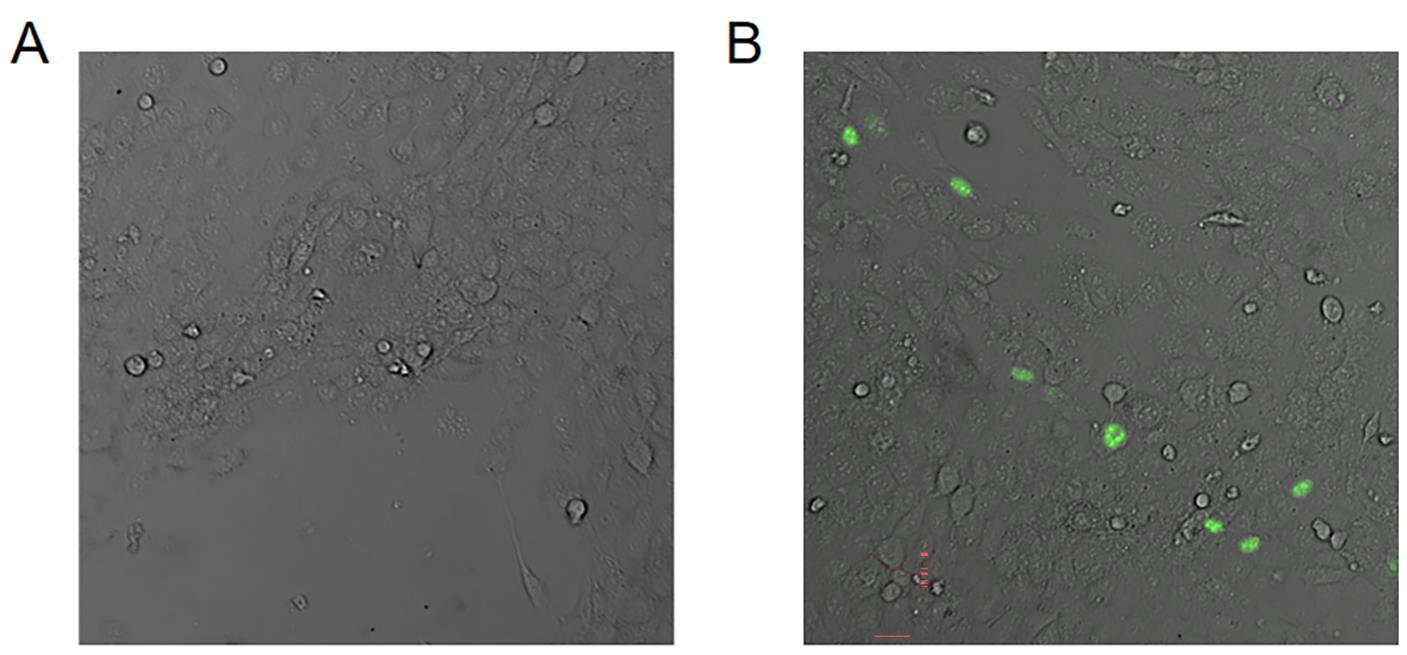
Figure 2. Representative images of GFP-expressing cells under fluorescent microscopy. Images of SAS (A) without or (B) with I-Scel transfection. Scale bar = 10 µm.Flow cytometry detection of GFP+ cells
Wash the infected cells (at least 1 × 106 cells) twice with 1× PBS and trypsinize into single cell suspension.
Wash again with 1× PBS, spin at 500 × g for 5 min, resuspend in 1 mL of 1× PBS, and force cells through a flow tube with cell strainer cap.
Set the gate of GFP/FITC-Area on flow cytometry to detect the proportion of GFP+ cells (Figure 3). The efficiency of Alt-EJ detection by flow cytometry is further validated by two classical inhibitors. One is TGFBR1 inhibitor LY2157299, which leads to an upregulation of Alt-EJ levels. The other is PARP1 inhibitor Olaparib, which leads to a down-regulation of Alt-EJ levels. As shown in Figure 3E, our improved reporter assay accurately reflected the effect of both inhibitors on the regulation of Alt-EJ levels.
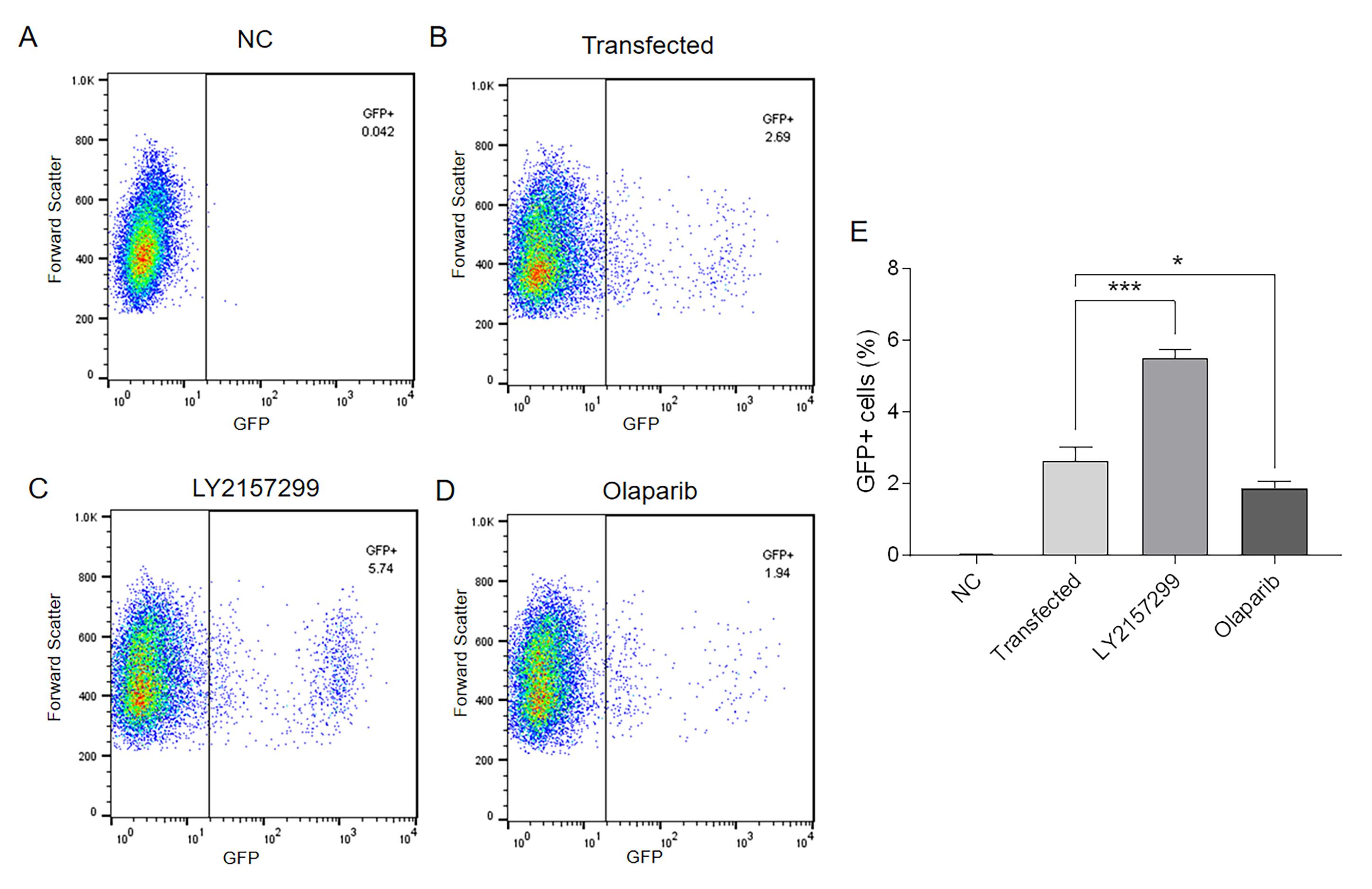
Figure 3. Flow cytometry detection of GFP+ SAS cells. (A) Negative control without I-SceI expression. (B) Control with I-SceI expression. (C) TGFBRI inhibition with LY2157299 (0.4 μmol/L for 48 h) to increase Alt-EJ. (D) PARP1 inhibition with Olaparib (10 μmol/L for 48 h) to decrease Alt-EJ. (E) Percentage of GFP-positive cells of SAS-EJ2-puro. NC: negative control; *P < 0.05; ***P < 0.001.
Recipes
1× PBS
Dissolve 137 mM of NaCl, 2.7 mM of KCl, 7.5 mM of Na2HPO4·12H2O, and 9.1 mM of KH2PO4 in 1 L ddH2O (pH 7.4).
SAS/HEK 293T medium
445 mL of DMEM supplemented with 50 mL of bovine growth serum and 5 mL of streptomycin-penicillin.
Data analysis
Flow cytometry
Gate cells of interest by setting up plots for forward scatter versus side scatter.
Using the forward scatter height versus forward scatter area excludes the doublets.
Untransfected cells were used as a negative control to identify the gate for detecting GFP expression negative cells. This gate was then used to evaluate the level of GFP expression in the target cells after transfection.
Acknowledgments
This study was supported by the Natural Science Foundation of China (82073007; Q. L.). The authors wish to acknowledge Prof. Jeremy Stark, Prof. Mary Helen Barcellos-Hoff, Prof. John Murnane, and Mr. Trevor Jones for their kind help and support with the results of this study. The protocol described here is adapted from previous studies (Gunn et al., 2012; Muraki et al., 2013).
Competing interests
The authors declare no competing interests.
References
- Bennardo, N., Cheng, A., Huang, N. and Stark, J. M. (2008). Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4(6): e1000110.
- Caracciolo, D., Riillo, C., Di Martino, M. T., Tagliaferri, P. and Tassone, P. (2021). Alternative Non-Homologous End-Joining: Error-Prone DNA Repair as Cancer's Achilles' Heel. Cancers (Basel) 13(6).
- Ceccaldi, R., Liu, J. C., Amunugama, R., Hajdu, I., Primack, B., Petalcorin, M. I., O'Connor, K. W., Konstantinopoulos, P. A., Elledge, S. J., Boulton, S. J., et al. (2015). Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 518(7538): 258-262.
- Fujii, S., Sobol, R. W. and Fuchs, R. P. (2022). Double-strand breaks: When DNA repair events accidentally meet. DNA Repair 112: 103303.
- Gunn, A. and Stark, J. M. (2012). I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol Biol 920: 379-391.
- Huang, R. and Zhou, P. K. (2021). DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct Target Ther 6(1): 254.
- Liu, Q., Lopez, K., Murnane, J., Humphrey, T. and Barcellos-Hoff, M. H. (2019). Misrepair in Context: TGFbeta Regulation of DNA Repair. Front Oncol 9: 799.
- Liu, Q., Palomero, L., Moore, J., Guix, I., Espin, R., Aytes, A., Mao, J. H., Paulovich, A. G., Whiteaker, J. R., Ivey, R. G., et al. (2021). Loss of TGFbeta signaling increases alternative end-joining DNA repair that sensitizes to genotoxic therapies across cancer types. Sci Transl Med 13(580).
- Liu, Q., Chen, G., Moore, J., Guix, I., Placantonakis, D. and Barcellos-Hoff, M. H. (2022). Exploiting Canonical TGFbeta Signaling in Cancer Treatment. Mol Cancer Ther 21(1): 16-24.
- Liu, Q., Ma, L., Jones, T., Palomero, L., Pujana, M. A., Martinez-Ruiz, H., Ha, P. K., Murnane, J., Cuartas, I., Seoane, J., et al. (2018). Subjugation of TGFbeta Signaling by Human Papilloma Virus in Head and Neck Squamous Cell Carcinoma Shifts DNA Repair from Homologous Recombination to Alternative End Joining. Clin Cancer Res 24(23): 6001-6014.
- Guix, I., Liu, Q., Pujana, M. A., Ha, P., Piulats, J., Linares, I., Guedea, F., Mao, J. H., Lazar, A., Chapman, J., et al. (2022). Validation of Anticorrelated TGFbeta Signaling and Alternative End-Joining DNA Repair Signatures that Predict Response to Genotoxic Cancer Therapy. Clin Cancer Res 28(7): 1372-1382.
- Mateos-Gomez, P. A., Gong, F., Nair, N., Miller, K. M., Lazzerini-Denchi, E. and Sfeir, A. (2015). Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 518(7538): 254-257.
- Muraki, K., Han, L., Miller, D. and Murnane, J. P. (2013). The role of ATM in the deficiency in nonhomologous end-joining near telomeres in a human cancer cell line. PLoS Genet 9(3): e1003386.
- Rahimian, E., Amini, A., Alikarami, F., Pezeshki, S. M. S., Saki, N. and Safa, M. (2020). DNA repair pathways as guardians of the genome: Therapeutic potential and possible prognostic role in hematologic neoplasms. DNA Repair 96: 102951.
- Valikhani, M., Rahimian, E., Ahmadi, S. E., Chegeni, R. and Safa, M. (2021). Involvement of classic and alternative non-homologous end joining pathways in hematologic malignancies: targeting strategies for treatment. Exp Hematol Oncol 10(1): 51.
- Wang, M., Chen, S. and Ao, D. (2021). Targeting DNA repair pathway in cancer: Mechanisms and clinical application. MedComm (2020) 2(4): 654-691.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zuo, N., Ma, L., Hu, W., Deng, Y., Wei, L. and Liu, Q. (2022). Detection of Alternative End-Joining in HNSC Cell Lines Using DNA Double-Strand Break Reporter Assays. Bio-protocol 12(17): e4506. DOI: 10.21769/BioProtoc.4506.
- Liu, Q., Palomero, L., Moore, J., Guix, I., Espin, R., Aytes, A., Mao, J. H., Paulovich, A. G., Whiteaker, J. R., Ivey, R. G., et al. (2021). Loss of TGFbeta signaling increases alternative end-joining DNA repair that sensitizes to genotoxic therapies across cancer types. Sci Transl Med 13(580).
Category
Cancer Biology > Genome instability & mutation > Genetics
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link