- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Microscopic Detection of DNA Synthesis in Early Mitosis at Repetitive lacO Sequences in Human Cells
Published: Vol 12, Iss 17, Sep 5, 2022 DOI: 10.21769/BioProtoc.4504 Views: 2637
Reviewed by: Gal HaimovichDavid PaulVaibhav B. Shah

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Quantification of Chromosomal Aberrations in Mammalian Cells
Inés Paniagua and Jacqueline J. L. Jacobs
Aug 20, 2023 2787 Views
Assay for Site-Specific Homologous Recombination Activity in Adherent Cells, Suspension Cells, and Tumor Tissues
Yuki Yoshino [...] Natsuko Chiba
Apr 5, 2025 2410 Views
Protocol for Quantifying γH2AX Foci in Irradiated Cells Using Immunofluorescence and Fiji Software
Lu Deng [...] Lingying Wu
Aug 20, 2025 2354 Views
Abstract
In the human cell cycle, complete replication of DNA is a fundamental process for the maintenance of genome integrity. Replication stress interfering with the progression of replication forks causes difficult-to-replicate regions to remain under-replicated until the onset of mitosis. In early mitosis, a homology-directed repair DNA synthesis, called mitotic DNA synthesis (MiDAS), is triggered to complete DNA replication. Here, we present a method to detect MiDAS in human U2OS 40-2-6 cells, in which repetitive lacO sequences integrated into the human chromosome evoke replication stress and concomitant incomplete replication of the lacO array. Immunostaining of BrdU and LacI proteins is applied for visualization of DNA synthesis in early mitosis and the lacO array, respectively. This protocol has been established to easily detect MiDAS at specific loci using only common immunostaining methods and may be optimized for the investigation of other difficult-to-replicate regions marked with site-specific binding proteins.
Keywords: DNA replicationBackground
A basic principle of the cell cycle is to ensure that mitosis follows the completion of DNA replication. During S phase, various replication stresses, such as DNA lesions, conflicts with transcription, and tightly bound protein complexes, interfere with DNA replication and cause a genomic instability related to various genetic diseases (Muñoz and Méndez, 2017). To prevent or minimize the deleterious consequences of replication stress, cells have evolved DNA damage response mechanisms, including DNA repair pathways (Yoshida and Fujita, 2021). Stalled replication forks activate checkpoint pathways that halt the cell cycle progression (Lemmens and Lindqvist, 2019; Mocanu and Chan, 2021). However, it has been revealed that upon mild replication stress that modestly slows down the replication fork, cells can enter the mitosis with some under-replicated DNA (Lukas et al., 2011; Bertolin et al., 2020; Lezaja and Altmeyer, 2021; Mocanu and Chan, 2021). Mild replication stress causes incomplete replication without activating G2/M checkpoint, especially at difficult-to-replicate regions such as chromosomal fragile sites, centromeres, and telomeres (Sarlós et al., 2017; Özer and Hickson, 2018; Lokanga et al., 2021). Such regions with under-replicated DNA activate mitotic DNA synthesis (MiDAS) in early mitosis to complete DNA replication, whereas some regions remain incomplete and will be resolved in the subsequent cell cycle (Minocherhomji et al., 2015; Özer and Hickson, 2018; Bertolin et al., 2020; Lezaja and Altmeyer, 2021).
MiDAS is a kind of homology-directed repair DNA synthesis that involves repair of stalled replication intermediates and POLD3 (an accessory subunit of polymerase δ)-dependent conservative DNA synthesis, analogous to break-induced replication (Kockler et al., 2021; Epum and Haber, 2022). Basic protocols for detection of MiDAS were originally developed by Hickson and colleagues (Minocherhomji et al., 2015; Bhowmick et al., 2016; Garribba et al., 2018). Briefly, cells are treated with low dose (0.4 μM) aphidicolin and RO-3306. Aphidicolin, a DNA polymerase inhibitor, slows down DNA replication and induces under-replication, while RO-3306, an inhibitor of cyclin-dependent kinase 1, arrests cells at G2 phase. Cells are then released into mitosis in the presence of a nucleoside analog ethynyl deoxyuridine (EdU) to label newly synthesized DNA. Following click reaction, MiDAS is visualized as EdU foci on prophase chromatin by fluorescence microscopy.
Here, we describe a detailed method for detection of MiDAS at under-replicated regions evoked by repetitive lacO sequences in human U2OS 40-2-6 cells (Figure 1). U2OS 40-2-6 cell, a U2OS-derived cell line carrying lacO array and estrogen receptor (ERT2)–fused LacI gene, was generated to investigate DNA damage response to replication stress induced by tight DNA–protein complexes on the human chromosome (Ishimoto et al., 2021). Treatment with 4-hydroxytamoxifen (4-OHT) induces rapid formation of lacO–LacI complexes that interfere with DNA replication. Notably, we also found that even in the absence of LacI, repetitive lacO sequences are intrinsically difficult to replicate, remaining under-replicated until mitosis and consequently triggering MiDAS. In the method presented here, nascent DNA is labeled by another nucleoside analog bromodeoxyuridine (BrdU), and lacO array is visualized by immunostaining of LacI proteins bound to the array. In our ER–LacI induction system, replication stress is induced at the specific chromosomal locus using the sequence-specific DNA-binding proteins. MiDAS at the specific locus can therefore be easily detected using common immunostaining methods, without fluorescent in situ hybridization (FISH) used in the protocol developed by Hickson and colleagues. This protocol may be optimized for other repetitive fragile sites where MiDAS functions, such as telomeres, centromeres, and ribosomal DNA arrays.
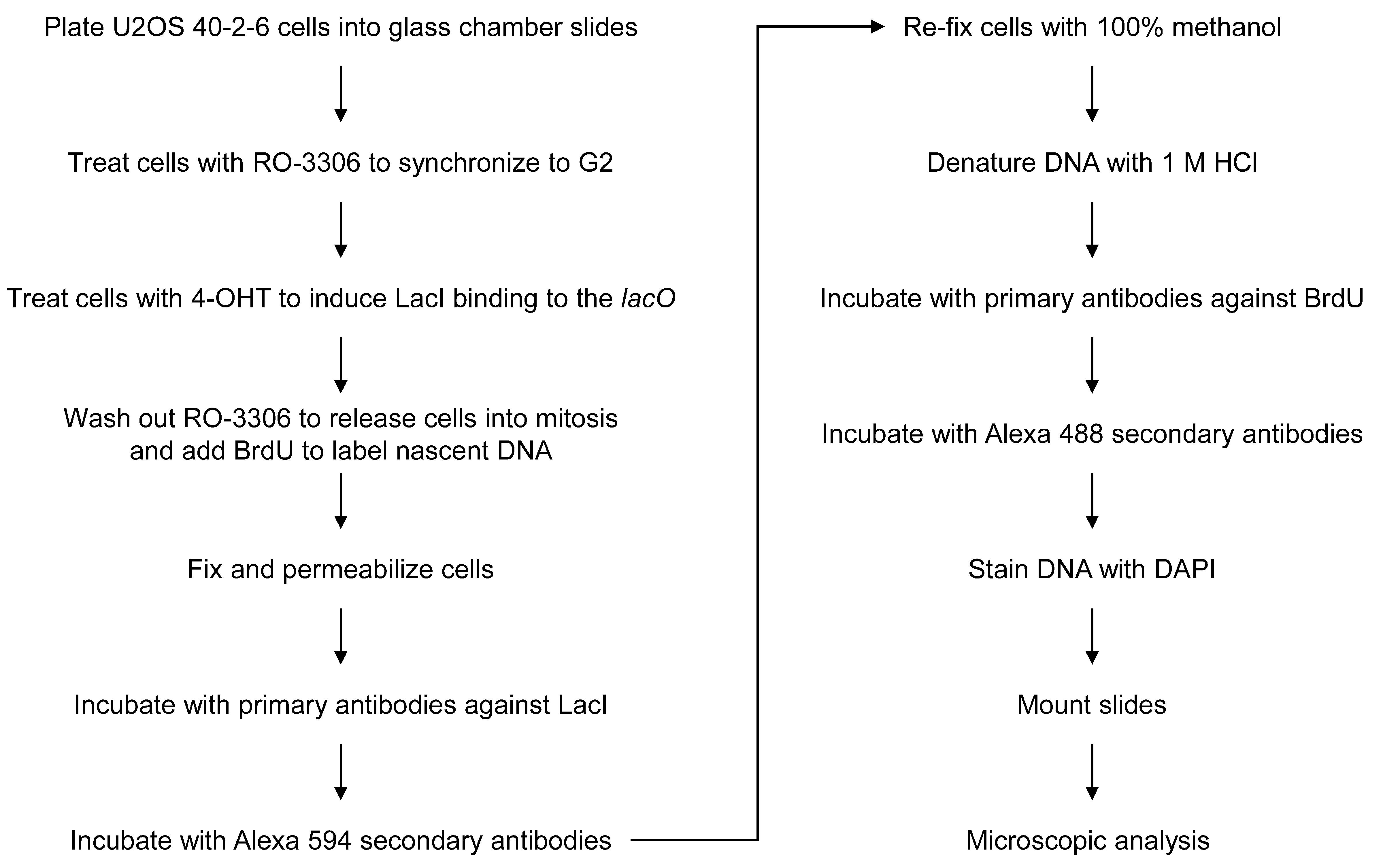
Figure 1. Flowchart of the assay for detection of mitotic DNA synthesis at the lacO array.
Materials and Reagents
Lab-Tek II, 4-well chamber glass slides (Thermo Fisher, Nunc, catalog number: 154526)
Chamber removal tool (included with chamber slides)
Glass Coplin jar, 5 slides type (AsOne, catalog number: 4-567-01)
Syringe filter unit, PES 0.22 μm (Merk Millipore, catalog number: SLGPR33RS)
Syringe filter unit, PVDF 0.45 μm (Merk Millipore, catalog number: SLHVR33RS)
Rectangular dish (Eiken Chemical, catalog number: AW2000)
Cotton swab
Paper towel (Nippon Paper Crecia, catalog number: 37105)
Glass coverslips, 24 × 60 mm (Matsunami Glass, catalog number: C024601)
Nail polish (clear topcoat)
Human cell line U2OS 40-2-6 (Ishimoto et al., 2021), a U2OS-derived cell line carrying lacO array and ERT2-LacI gene generated from U2OS 2-6-3 cells (Janicki et al., 2004). U2OS 40-2-6 cells, and the original U2OS 2-6-3 cells, contain an approximately 200-copy array of 256 lacO repeats (approximately 50,000 copies of lacO sequences in total). The array is stably integrated into the chromosomal locus 1p36 in chromosome 1.
Dulbecco’s modified Eagle’s medium (D-MEM) (high glucose with L-glutamine) (e.g., Wako, catalog number: 043-30085)
Fetal bovine serum (FBS) (e.g., Nichirei, catalog number: 175012)
RO-3306 (e.g., Sigma-Aldrich, catalog number: SML0569)
4-Hydroxytamoxifen (4-OHT) (e.g., Abcam, catalog number: ab141943)
Ethanol (e.g., Nacalai Tesque, catalog number: 14713-53)
10× phosphate-buffered saline (PBS) (e.g., Wako, catalog number: 048-29805), diluted to 1×
Bromodeoxyuridine (BrdU) (e.g., Sigma-Aldrich, catalog number: B5002)
Saline (e.g., Otsuka Normal Saline, Otsuka Pharmaceutical)
PIPES (e.g., Dojindo, catalog number: 347-02224)
0.1 M EGTA-2Na solution (e.g., Nacalai Tesque, catalog number: 37346-05)
Triton X-100 (e.g., Wako, catalog number: 169-21105)
MgCl2 (e.g., Wako, Catalog number: 133-00165)
37% formaldehyde solution (e.g., Nacalai Tesque, catalog number: 16223-55)
Rabbit anti-LacI antibody, homemade (Ishimoto et al., 2021)
Thimerosal (e.g., Nacalai Tesque, Catalog number: 21624-74), dissolved with water to 10% (w/v)
Alexa 594 donkey anti-Rabbit IgG (Molecular Probes, catalog number: A21207)
Methanol (e.g., Nacalai Tesque, catalog number: 21915-93)
HCl (e.g., Wako, catalog number: 080-01066), diluted to 1 M
Mouse anti-BrdU antibody, clone: 3D4 (BD Pharmingen, catalog number: 555627)
Alexa 488 goat anti-Mouse IgG (Molecular Probes, catalog number: A11029)
4’, 6-diamidino-2-phenylindole (DAPI) (e.g., Wako, catalog number: 043-18804)
Fluoro-KEEPER antifade reagent (Nacalai Tesque, catalog number: 12593-64)
RO-3306 stock solution (see Recipes)
4-OHT stock solution (see Recipes)
BrdU stock solution (see Recipes)
PTEMF buffer (see Recipes)
FBS for antibody working solution (see Recipes)
Antibody working solutions (see Recipes)
DAPI stock solution (see Recipes)
Note: In our lab, we routinely use DAPI, Alexa 488, and Alexa 594 for triple staining of DNA and two interest proteins. Other combinations of fluorophores may be possible.
Equipment
Cell culture incubator (ASTEC, model: SCA-165D)
Water aspirator (e.g., FLINN scientific, model: AP1136)
Widefield fluorescence microscope equipped with a cooled CCD camera, an LED illumination source, and filters to image Alexa 488, Alexa 594, and DAPI (e.g., Keyence, BZ-X700). Similar conventional fluorescence microscopes would be applicable.
40× objective lens [e.g., Nikon, CFI (chromatic aberration free infinity) Plan Apo λ 40×/0.95 NA]. Higher magnification objectives, such as a 63× lens, may also be applicable.
Software
Microscope operating software (e.g., Keyence, the BZ-X viewer software)
Procedure
Cell synchronization and induction of LacI binding
Plate U2OS 40-2-6 cells at 4 × 104 cells in 0.5 mL of D-MEM medium supplemented with 8% FBS for each well of a 4-well chamber slide, so that the cells are 50%–60% confluent the next day.
Incubate cells in a cell culture incubator with 5% CO2 at 37 °C overnight.
Add RO-3306 to a final concentration of 7 μM to the D-MEM medium supplemented with 8% FBS in each well.
(Optional) For induction of LacI binding in S phase, add 4-OHT to a final concentration of 1 μM.
Note: LacI binding in S phase induces additional replication stress; however, it does not increase a frequency of MiDAS, probably because of the limited capacity for MiDAS (Ishimoto et al., 2021).
Incubate cells with RO-3306 at 37 °C for 20 h to synchronize cells at late G2 phase.
After 16 h incubation, to induce LacI binding in G2 phase, add 4-OHT to a final concentration of 1 μM (4 h treatment with 4-OHT in the presence of RO-3306) for the remaining 4 h of incubation with RO-3306.
Release into mitosis and labeling with BrdU
After 20 h incubation, cells are now in late G2 phase.
Wash cells three times with 1 mL of 1× PBS, pre-warmed to 37 °C, to remove RO-3306.
Add 0.5 mL of pre-warmed D-MEM medium supplemented with 8% FBS.
Simultaneously, add BrdU to a final concentration of 10 μM to label nascent DNA.
Incubate cells at 37 °C for 20 min, allowing cells to enter prophase.
Note: Be quick with washing and treating cells to limit exposure to room temperature (RT), which may disturb cell cycle progression. Pre-warming PBS and D-MEM to 37 °C is also critical for rapid and synchronous entry into mitosis.
Cell fixation, permeabilization, and immunostaining of LacI
Gently remove growth medium with a water aspirator attached with a yellow tip (200 μL).
Carefully add 1 mL of ice-cold PBS to each well.
Gently remove PBS, and carefully add 1 mL of PTEMF buffer for simultaneous fixation and permeabilization of the cells.
Incubate at RT for 20 min.
Aspirate PTEMF buffer and disassemble chambers from the glass slide. Be careful not to dry out cells during disassembly.
Note: Glass slides could be handled by hand, but tweezers should be used to remove and transfer the slides from the solution in Coplin jar.
Wash slides by immersing them in Coplin jar containing 50 mL of ice-cold PBS, three times for 5 min each.
Place slides in a rectangular dish facing up and remove excess PBS in the hydrophobic area around the wells with a cotton swab. Be quick not to dry out cells.
Add 95 μL of primary anti-LacI antibody solution (diluted 1:500, see Recipes) to each well.
Close the lid of rectangular dish and incubate at RT for 1 h.
Gently drain the antibody solution by tilting slides on paper towel.
Wash slides with ice-cold PBS in Coplin jar three times for 5 min each.
Place slides in a rectangular dish facing up and remove excess PBS with a cotton swab. Be quick not to dry out cells.
Add 95 μL of Alexa 594 secondary anti-rabbit antibody solution (diluted 1:1,000, see Recipes) to each well.
Close the rectangular dish and incubate at RT for 1 h.
From now on, shield the samples from light to avoid photobleaching of fluorescent dyes.
Re-fixation, DNA denaturation, and immunostaining of BrdU
Gently drain the antibody solution by tilting slides on paper towel.
Wash slides with 50 mL of ice-cold PBS in Coplin jar, three times for 5 min each.
Refix samples by immersing slides in Coplin jar containing 100% methanol (pre-chilled in -30 °C freezer overnight) for 10 min on the bench (at RT).
Wash slides with 50 mL of ice-cold PBS in Coplin jar, three times for 5 min each.
To denature DNA, immerse slides in Coplin jar containing 50 mL of 1M HCl for 15 min at RT.
Note: DNA denaturation is required for immunostaining of BrdU; however, too much treatment with HCl may cause weak DAPI staining at step E3.
Wash slides with ice-cold PBS in Coplin jar, three times for 5 min each.
Place slides in a rectangular dish facing up and remove excess PBS with a cotton swab. Be quick not to dry out cells.
Add 95 μL of primary anti-BrdU antibody solution (diluted 1:200, see Recipes) to each well.
Close the rectangular dish and incubate at RT for 1 h.
Gently drain the antibody solution by tilting slides on paper towel.
Wash slides with ice-cold PBS in Coplin jar, three times for 5 min each.
Place slides in a rectangular dish facing up and remove excess PBS with a cotton swab. Be quick not to dry out cells.
Add 95 μL of Alexa 488 secondary anti-mouse antibody solution (diluted 1:1,000, see Recipes) to each well.
Close the rectangular dish and incubate at RT for 1 h.
DAPI staining and mounting
Gently drain the antibody solution by tilting slides on paper towel.
Wash slides with 50 mL of ice-cold PBS in Coplin jar, three times for 5 min each.
Immerse slides in Coplin jar containing DAPI working solution (0.2 μg/mL, protected from light) in PBS for 15 min at RT.
Wash slides with 50 mL of ice-cold PBS in Coplin jar, three times for 5 min each.
Place slides on a paper towel facing up and remove excess PBS by cotton swab, taking care to prevent cells from drying out.
Add one drop of Fluoro-KEEPER antifade reagent to each well of the slide.
Mount with a glass coverslip, ensuring to avoid air bubbles.
Wipe off excess Fluoro-KEEPER antifade reagent from the edge of the coverslip.
Seal with nail polish at the edge of the coverslip and let it out to dry and cure at RT for at least 2 h or overnight.
Proceed to microscopy or store the slide at 4 °C for 1–2 months, keeping protected from light.
Image acquisition
Image the slide using a fluorescence microscope.
Find an area including cells in prophase/prometaphase using the DAPI channel and focus on LacI foci using the Alexa 594 channel.
Acquire images for each channel (DAPI, Alexa 488, and Alexa 594) consecutively at the same field position without changing the focus (Figure 2).
To evaluate the induction of MiDAS at lacO arrays, calculate colocalization frequencies of BrdU foci with lacO arrays. The numbers of prophase/prometaphase nuclei with a LacI focus and frequencies of nuclei in which BrdU signal forms a focus on the LacI focus are manually scored. Although prophase/prometaphase cells are enriched in this condition, BrdU foci colocalized with lacO foci can be observed in prophase/prometaphase and metaphase cells. In contrast, BrdU incorporation is not detected in G2-arrested cells (not released from RO-3306), as previously reported (Ishimoto et al., 2021).
Note: These images should be compared with the control assays without BrdU labeling (Figure 2, bottom) to confirm the specificity of the BrdU signals.
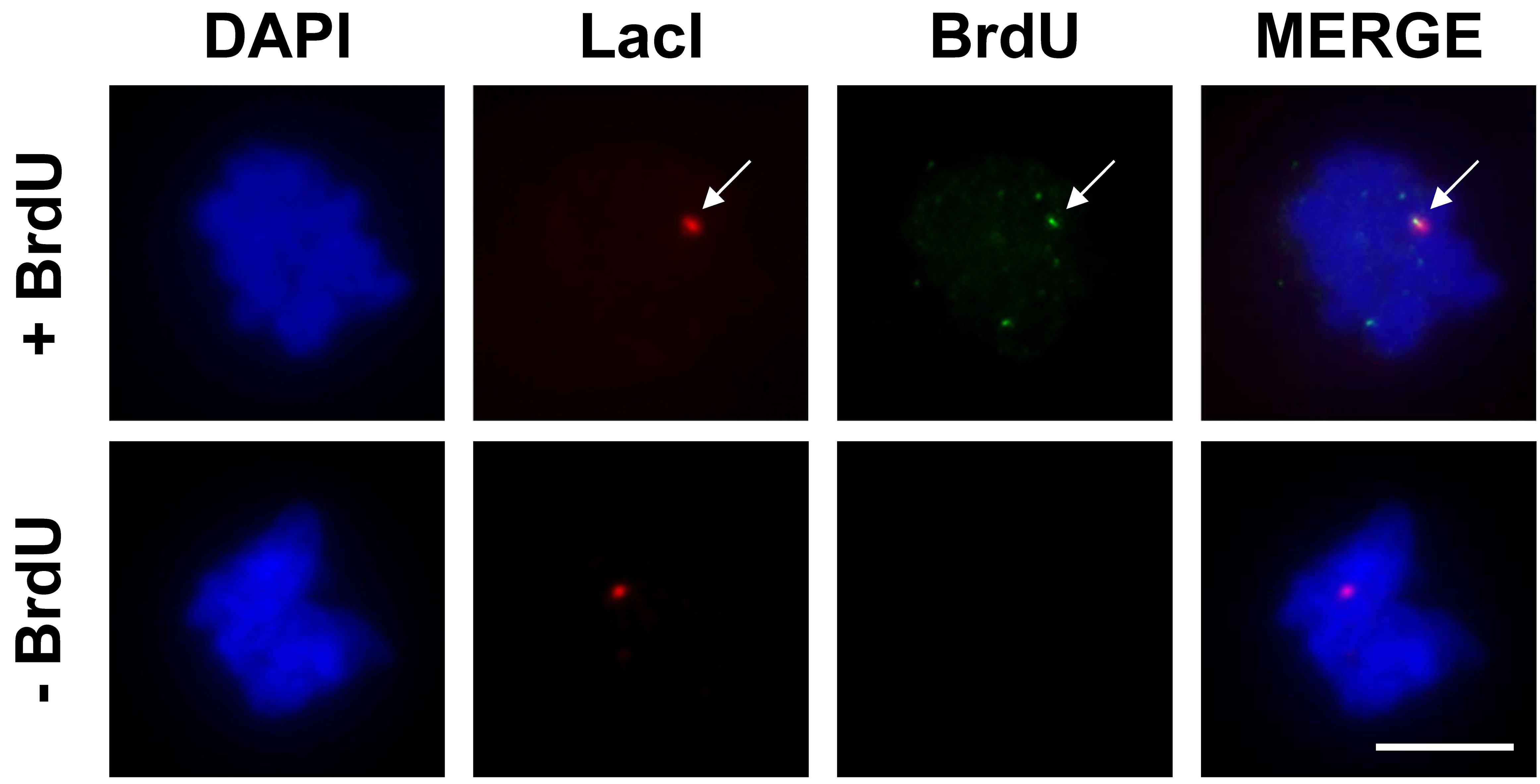
Figure 2. Representative images of mitotic DNA synthesis at the lacO array. The colocalization of LacI and BrdU is indicated by the white arrow. Scale bar = 10 μm.
Recipes
RO-3306 stock solution
Dissolve RO-3306 in DMSO to a stock concentration of 1 mM and store aliquots at -80 °C. Pre-warm to 37 °C before use.
4-OHT stock solution
Dissolve 4-OHT in 100% ethanol to a stock concentration of 125 μM and store aliquots at -30 °C. Pre-warm to 37 °C before use.
BrdU stock solution
Dissolve BrdU in saline to a stock concentration of 10 mg/mL (32.5 mM) and store aliquots at -80 °C.
PTEMF buffer
20 mM PIPES, pH 6.8
10 mM EGTA
0.2% Triton X-100
1 mM MgCl2
4% formaldehyde
Note: Prepare a solution without formaldehyde, filtrate with 0.22 μm filter, and store at 4 °C. Add formaldehyde freshly for each experiment.
FBS for antibody working solution
Add thimerosal to FBS to a concentration of 0.1% and filtrate with 0.45 μm filter. Store at 4 °C or at -30 °C for long-term storage.
Antibody working solutions
Dilute antibodies in 1× PBS supplemented with 10% FBS. Freshly prepare for each experiment.
Detection of LacI
Primary antibody: Rabbit anti-LacI, 1:500
Secondary antibody: Alexa 594 anti-Rabbit, 1:1,000
Detection of BrdU
Primary antibody: Mouse anti-BrdU, 1:200
Secondary antibody: Alexa 488 anti-Mouse, 1:1,000
DAPI stock solution
Dissolve DAPI in water to a stock concentration of 1 mg/mL, protected from light. Store at -30 °C.
Acknowledgments
This work was supported by grants from the Japan Society for the Promotion of Science (KAKENHI JP15K18478 and JP22H02603), the Fukuoka Foundation for Sound Health Cancer Research Fund, and the Mochida Memorial Foundation for Medical and Pharmaceutical Research. This protocol was adapted from our previous work (Ishimoto et al., 2021). Basic methods for detection of MiDAS were originally developed by the laboratory of Dr. Ian Hickson (Minocherhomji et al., 2015; Bhowmick et al., 2016)
Competing interests
The authors declare no conflicting interests.
References
- Bertolin, A. P., Hoffmann, J. S. and Gottifredi, V. (2020). Under-Replicated DNA: The Byproduct of Large Genomes? Cancers (Basel) 12(10): 2764.
- Bhowmick, R., Minocherhomji, S. and Hickson, I. D. (2016). RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell 64(6): 1117-1126.
- Epum, E. A. and Haber, J. E. (2022). DNA replication: the recombination connection. Trends Cell Biol 32(1): 45-57.
- Garribba, L., Wu, W., Ozer, O., Bhowmick, R., Hickson, I. D. and Liu, Y. (2018). Inducing and Detecting Mitotic DNA Synthesis at Difficult-to-Replicate Loci. Methods Enzymol 601: 45-58.
- Ishimoto, R., Tsuzuki, Y., Matsumura, T., Kurashige, S., Enokitani, K., Narimatsu, K., Higa, M., Sugimoto, N., Yoshida, K. and Fujita, M. (2021). SLX4-XPF mediates DNA damage responses to replication stress induced by DNA-protein interactions. J Cell Biol 220(1): e202003148.
- Janicki, S. M., Tsukamoto, T., Salghetti, S. E., Tansey, W. P., Sachidanandam, R., Prasanth, K. V., Ried, T., Shav-Tal, Y., Bertrand, E., Singer, R. H., et al. (2004). From silencing to gene expression: real-time analysis in single cells. Cell 116(5): 683-698.
- Kockler, Z. W., Osia, B., Lee, R., Musmaker, K. and Malkova, A. (2021). Repair of DNA Breaks by Break-Induced Replication. Annu Rev Biochem 90: 165-191.
- Lemmens, B. and Lindqvist, A. (2019). DNA replication and mitotic entry: A brake model for cell cycle progression. J Cell Biol 218(12): 3892-3902.
- Lezaja, A. and Altmeyer, M. (2021). Dealing with DNA lesions: When one cell cycle is not enough. Curr Opin Cell Biol 70: 27-36.
- Lokanga, R. A., Kumari, D. and Usdin, K. (2021). Common Threads: Aphidicolin-Inducible and Folate-Sensitive Fragile Sites in the Human Genome. Front Genet 12: 708860.
- Lukas, C., Savic, V., Bekker-Jensen, S., Doil, C., Neumann, B., Pedersen, R. S., Grofte, M., Chan, K. L., Hickson, I. D., Bartek, J., et al. (2011). 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13(3): 243-253.
- Minocherhomji, S., Ying, S., Bjerregaard, V. A., Bursomanno, S., Aleliunaite, A., Wu, W., Mankouri, H. W., Shen, H., Liu, Y. and Hickson, I. D. (2015). Replication stress activates DNA repair synthesis in mitosis. Nature 528(7581): 286-290.
- Mocanu, C. and Chan, K. L. (2021). Mind the replication gap. R Soc Open Sci 8(6): 201932.
- Muñoz, S. and Méndez, J. (2017). DNA replication stress: from molecular mechanisms to human disease. Chromosoma 126(1): 1-15.
- Özer, Ö. and Hickson, I. D. (2018). Pathways for maintenance of telomeres and common fragile sites during DNA replication stress. Open Biol 8(4): 180018.
- Sarlós, K., Biebricher, A., Petermann, E. J. G., Wuite, G. J. L. and Hickson, I. D. (2017). Knotty Problems during Mitosis: Mechanistic Insight into the Processing of Ultrafine DNA Bridges in Anaphase. Cold Spring Harb Symp Quant Biol 82: 187-195.
- Yoshida, K. and Fujita, M. (2021). DNA damage responses that enhance resilience to replication stress. Cell Mol Life Sci 78(21-22): 6763-6773.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Yoshida, K., Ishimoto, R. and Fujita, M. (2022). Microscopic Detection of DNA Synthesis in Early Mitosis at Repetitive lacO Sequences in Human Cells. Bio-protocol 12(17): e4504. DOI: 10.21769/BioProtoc.4504.
- Ishimoto, R., Tsuzuki, Y., Matsumura, T., Kurashige, S., Enokitani, K., Narimatsu, K., Higa, M., Sugimoto, N., Yoshida, K. and Fujita, M. (2021). SLX4-XPF mediates DNA damage responses to replication stress induced by DNA-protein interactions. J Cell Biol 220(1): e202003148.
Category
Cancer Biology > Genome instability & mutation > Cell biology assays > DNA structure and alterations
Cancer Biology > General technique > Cell biology assays > Cell cycle
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link