- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Cryptococcus neoformans Virulence Assay Using a Galleria mellonella Larvae Model System
(*contributed equally to this work) Published: Vol 12, Iss 15, Aug 5, 2022 DOI: 10.21769/BioProtoc.4480 Views: 3080
Reviewed by: Kristin L. ShinglerLoredana ScalschiKrishna Saharan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In Vitro Hyphal Branching Assay Using Rhizophagus irregularis
Takaya Tominaga and Hironori Kaminaka
Aug 20, 2024 2338 Views

Silencing Arbuscular Mycorrhizal Fungal Gene Using Chitosan Nanoparticle-Mediated dsRNA Delivery System
Chumei Yan [...] Xianan Xie
Jun 5, 2025 2645 Views

A Reliable In Planta Inoculation and Antifungal Screening Protocol for Rhizoctonia solani-Induced Sheath Blight in Rice
Alinaj Yasin [...] Palash Deb Nath
Nov 5, 2025 1582 Views
Abstract
Cryptococcus neoformans is a human pathogenic fungus that can cause pulmonary infections and meningitis in both immunocompromised and otherwise healthy individuals. Limited treatment options and a high mortality rate underlie the necessity for extensive research of the virulence of C. neoformans. Here we describe a detailed protocol for using the Galleria mellonella (Greater Wax Moth) larvae as a model organism for the virulence analysis of the cryptococcal infections. This protocol describes in detail the evaluation of G. mellonella larvae viability and the alternatives for troubleshooting the infection procedure. This protocol can be easily modified to study different inocula or fungal species, or the effects of a drug or antifungal agent on fungal disease within the larvae. We describe modified alternative versions of the protocol that allow using G. mellonella to study fungal diseases with different inocula and at different temperatures.
Keywords: Cryptococcus neoformansBackground
Cryptococcus neoformans is a basidiomycetous polysaccharide-encapsulated fungus, pathogenic for multiple species (Kwon-Chung et al., 2014). In humans, C. neoformans can cause serious infections, primarily in immunocompromised patients with clinical manifestations of pneumonia and meningitis (Lui et al., 2006; Kwon-Chung et al., 2014). Due to high mortality, lack of available vaccines, limited treatment options, and increasing resistance to the fluconazole—a drug commonly used for treatment of cryptococcosis—it is estimated that the total number of cryptococcosis-related deaths exceeds 180,000 annually worldwide (Cogliati, 2013; Perfect and Bicanic, 2015). Cryptococcus spp. have several characteristic virulence factors that are essential for establishing infection and providing defense mechanisms against the host immune system. Important cryptococcal virulence factors include the polysaccharide capsule, melanin, and formation of titan cells (Zaragoza, 2019). Researchers have developed several methods to stimulate the production of those virulence factors in culture conditions; while this allows for precise functional analysis, it is not sufficient for a more holistic understanding of virulence (Casadevall et al., 2000; Liu et al., 2008; Crabtree et al., 2012; Almeida et al., 2015).
The study of virulence usually requires the use of an animal model organism. Animal testing plays an essential role in the medical and biomedical field of research. Unfortunately, research performed on mice, rats, and other mammals comes with serious limitations, including medium to high costs, requirement for specialized animal testing facilities, and concerns about the use of vertebrate animals in research. Additionally, mouse and mammalian studies can take several months depending on the virulence of the microbe and disease progression. To avoid those problems, many researchers performed studies, including virulence assays, on invertebrate models like the fruit fly Drosophila melanogaster or the larvae of the Greater wax moth Galleria mellonella. G. mellonella has been shown to be a useful model organism to test pathogenicity of multiple fungal species, including C. neoformans, which was first used in a G. mellonella model by the Mylonakis group in 2005 (Reeves et al., 2004; Mylonakis et al., 2005; Pereira et al., 2018; Maurer et al., 2019). The advantage of G. mellonella as a model host is its flexibility for changes in the infection protocol that allow the study of variables not possible in vertebrate and invertebrate models, such as host temperature. In contrast to other invertebrate model systems, infected larvae of G. mellonella can be incubated at room temperature, 30 °C, and 37 °C (Mylonakis et al., 2005). This temperature range is important because it allows experiments at mammalian temperatures. G. mellonella larvae are between 2–3 cm in length, an easy size to handle and manipulate. In addition, since G. mellonella are relatively easy to infect and store in high numbers, this model enables researchers to perform virulence or survival screens of many different conditions, strains, or mutants, before narrowing those down to use in a resource and time-intensive mouse model (Mylonakis et al., 2005; Firacative et al., 2020).
G. mellonella are used as models for C. neoformans infections, as several of the virulence factors important for C. neoformans infections in mammalian hosts also play comparable roles in G. mellonella. These include laccase, capsule, and the production of titan cells, which allow the fungus to evade immune clearance (Mylonakis et al., 2005; García-Rodas et al., 2011; Eisenman et al., 2014). Additionally, C. neoformans undergoes similar interactions with the immune systems of G. mellonella and mammals (Browne et al., 2013; Trevijano-Contador and Zaragoza, 2018; Stączek et al., 2020; Smith and Casadevall, 2021). C. neoformans is phagocytosed by both insect hemocytes (immune cells) and mammalian macrophages (Browne et al., 2013; Pereira et al., 2018). The fungus is also encapsulated within immune nodules of G. mellonella, which are aggregates of insect immune cells that neutralize the infection, comparable to the granulomas that form around C. neoformans within the mammalian lung. Here we present a standardized protocol for the virulence survival assay of C. neoformans performed in G. mellonella larvae.
Materials and Reagents
Snap-Cap 14 mL culture tubes (Falcon, catalog number: 352059)
Sterile inoculation loop (Fisherbrand, catalog number: 22-363-605)
Pipette tips 200 µL, 1,000 µL (USA Scientific TipOne, catalog numbers: 1111-0006, 1126-7810)
Insulin syringe with 0.400 (27G) needle (BD, catalog number: 329412)
10 cm Petri dish, one per each testing group (Falcon, catalog number:351029)
Nitrile Gloves (Halyard, catalog number: 55082)
1.5 mL Eppendorf tubes (SealRite, catalog number: 1615-5500)
Galleria mellonella larvae (https://www.waxworms.net/, Vanderhorst Wholesale, St. Marys, Ohio, USA)
Cryptococcus neoformans strain H99 (serotype A)
1× DPBS (Gibco, catalog number: 14190-144)
BD Bacto Peptone (Gibco, catalog number: 211677)
BD Bacto Yeast Extract (Gibco, catalog number: 212750)
Difco YPD Broth (Gibco, catalog number: 242820)
Small Polystyrene Weigh Boats (Heathrow Scientific, catalog number: HS1420A)
Glucose (Sigma, catalog number: 1002789701)
YPD liquid media (see Recipes)
Equipment
Hemocytometer (Hausser Scientific, catalog number: 3520)
Pipettes 2–20 µL, 20–200 µL, 100–1,000 µL
Microbiological incubator with a culture tube rotator
Microscope (Olympus, model: BX40)
Autoclave
Syringe stepper (Dymax STEPPER Repetitive Pipette, model: 4001-010)
Benchtop centrifuge (Spectrafuge 24D)
Software
GraphPad Prism software (https://www.graphpad.com/)
Procedure
Selection of larvae with desirable mass
Select and separate G. mellonella larvae and prepare Petri dishes, one for each tested group. Larvae are always handled while wearing nitrile gloves to reduce the risk of contamination.
Use a digital scale to select larvae with a body mass between 100 and 200 mg. Larvae are weighed individually in small polystyrene weigh boats before being transferred into Petri dishes, with up to 15 larvae in each Petri dish.
Note: Larvae should be relatively firm to the touch and not exhibit signs of illness, such as dark melanization throughout the body or reduced movement (Figure 1A). There is normal variability in the cuticle pigmentation in healthy larvae. Ideally, larvae will be sorted at least 16–24 h prior to infection to allow acclimatization at room temperature following arrival.
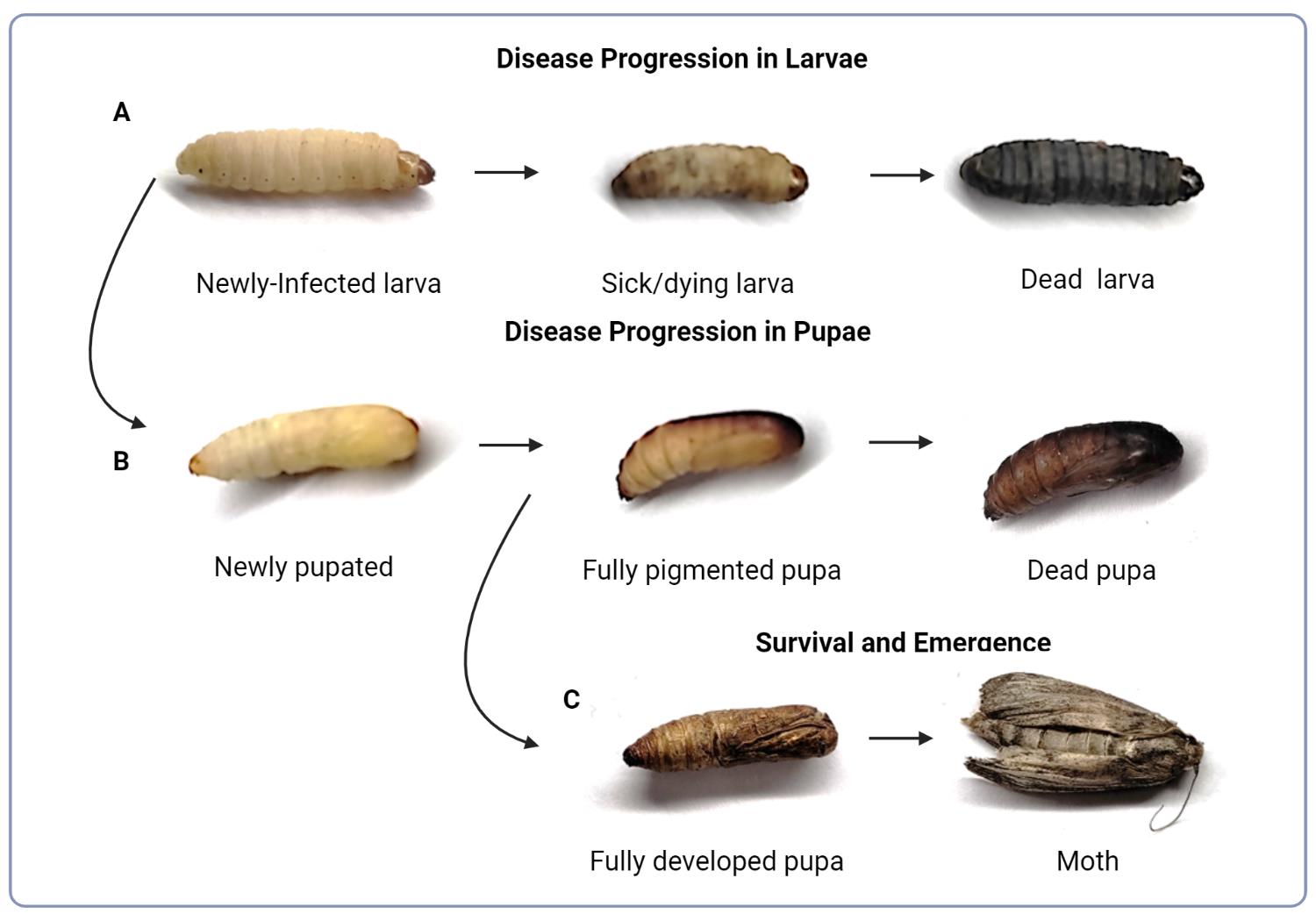
Figure 1. Evaluation of G. mellonella fitness. (A) Progression of cryptococcal infection in a larva resulting in discoloration and death. (B) Transition of infected larva into the pupa and progression of cryptococcal infection resulting in death. (C) Completed development of the pupa and metamorphosis into moth, which typically occurs at or beyond day 14 under the experimental lab conditions described.
Preparation of fungal cell inoculum
Approximately 48 h prior to the infection, inoculate 5 mL of YPD media with the desired strain of C. neoformans in a 14 mL Snap-Cap culture tube and incubate on the cell culture rotor at 30 °C, 35 rpm. Alternatively, C. neoformans can be grown in 1 mL of YPD media overnight (16–24 h) at 30 °C, 35 rpm.
Transfer 1 mL of stationary-phase culture to 1.5 mL Eppendorf tubes and pellet fungal strain by centrifuging tubes for 4 min at 2,500 × g in the tabletop centrifuge.
Wash your samples twice with DPBS by discarding supernatant and resuspending the pellet in 1 mL of sterile DPBS each time.
Prepare a 1:100 cell dilution and establish cell concentration using a hemocytometer.
According to this protocol, diluting cells to this concentration will result in 105 cells injected per larvae; if a different inoculum is desired, adjust the cell dilution appropriately. To prepare samples for injecting 105 cells per larvae, dilute your washed cells with DPBS to the concentration of 107 cells/mL.
Infection of larvae
Calibrate the stepper device and set to dispense 10 µL of liquid prior to the experiment, unless another volume is needed or desired.
Draw inoculum into a 1 mL tuberculin syringe.
Invert and flick the syringe until air bubbles rise to the top and air can be expelled, leaving only the prepared inoculum.
Load the syringe into the stepper device, making sure the syringe is firmly in place, the syringe’s plunger rests against the stepper’s pusher, and the hilt of the syringe rests on syringe clip to prevent slipping and inaccurate dispensing of inoculum.
Test that the stepper is accurately dispensing the inoculum when the dispense button is pressed.
Pick up a pre-sorted G. mellonella larva with the non-dominant hand. Hold the larva firmly with thumb and index fingers to prevent movement. Fingers should be placed towards the head and thorax to keep the abdomen open. Be careful not to stick the finger with the syringe needle.
Inject the needle into the last left proleg (Figure 2).
It is important to be consistent with which proleg is injected to reduce variability in the infection. The needle should enter midway through the larva.
Remove larvae carefully from the needle and place it into a Petri dish.
If excessive hemolymph (as indicated by yellow fluid) leaks from the injection site, larval death may occur. If problems with hemolymph leaking persist, leaving the needle in the larvae for 15 s before removal may reduce the amount of hemolymph lost. If there are concerns about dried hemolymph on the Petri dish following injection, hemolymph can be cleaned by temporarily removing the larvae, wiping the Petri dish with 70% ethanol, and drying with a paper towel.
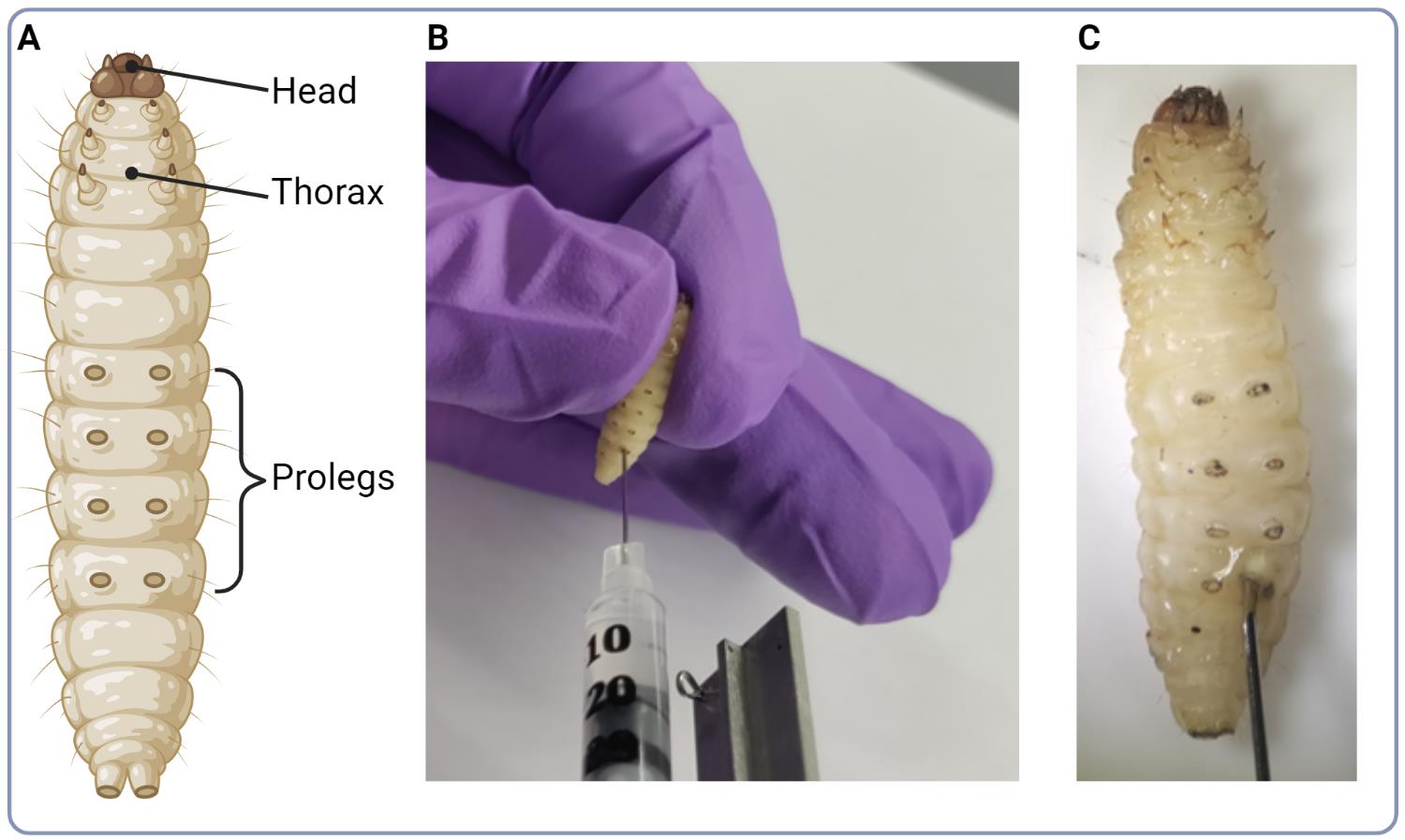
Figure 2. Infection of larvae. (A) Illustration of G. mellonella larva model with indication of major anatomical parts. (B) Photo illustrating the injection site of C. neoformans cell suspension into the base of larva’s proleg. (C) Close up photo of the injection site.
Incubation and maintenance of the tested larvae
After finishing the series of injections, transfer Petri dishes with larvae to the location with desired temperature for the analysis (room temperature or 30 °C).
Perform evaluation of larvae variability every day by checking for changes in color of each larva and by gently poking the larva using the pointy end of a pipette tip.
If survival of pupae is also assayed, check pupal survival by gently pressing down on pupae with the base of the pipette tip. Lack of movement following poking indicates death. Change of body color in deceased G. mellonella larvae progress from an initial creamy color to grey, brown, and black (Figure 1A). Healthy pupae start off white and yellow, and naturally become a light to medium brown color, whereas dead pupae show a dark brown to black color (Figure 1B).
Data analysis
To analyze and visualize differences in G. mellonella, upload the data into the Prism GraphPad software using the survival table format. Plot the data as a percentage of survival in relation to the time (days). An example of how the survival data is collected and how tables should be formatted in the Prism GraphPad format is found in Figure 3A and 3B, where 0 represents a censored event, where survival data can no longer be read for the organism (i.e., the organism survives to the endpoint of the experiment or is lost), 1 represents death of an organism, and blank cells indicate no event occurred (no larvae died). Each event (death or censoring) requires a new line. An example output of this data on Prism GraphPad is found in Figure 3C.
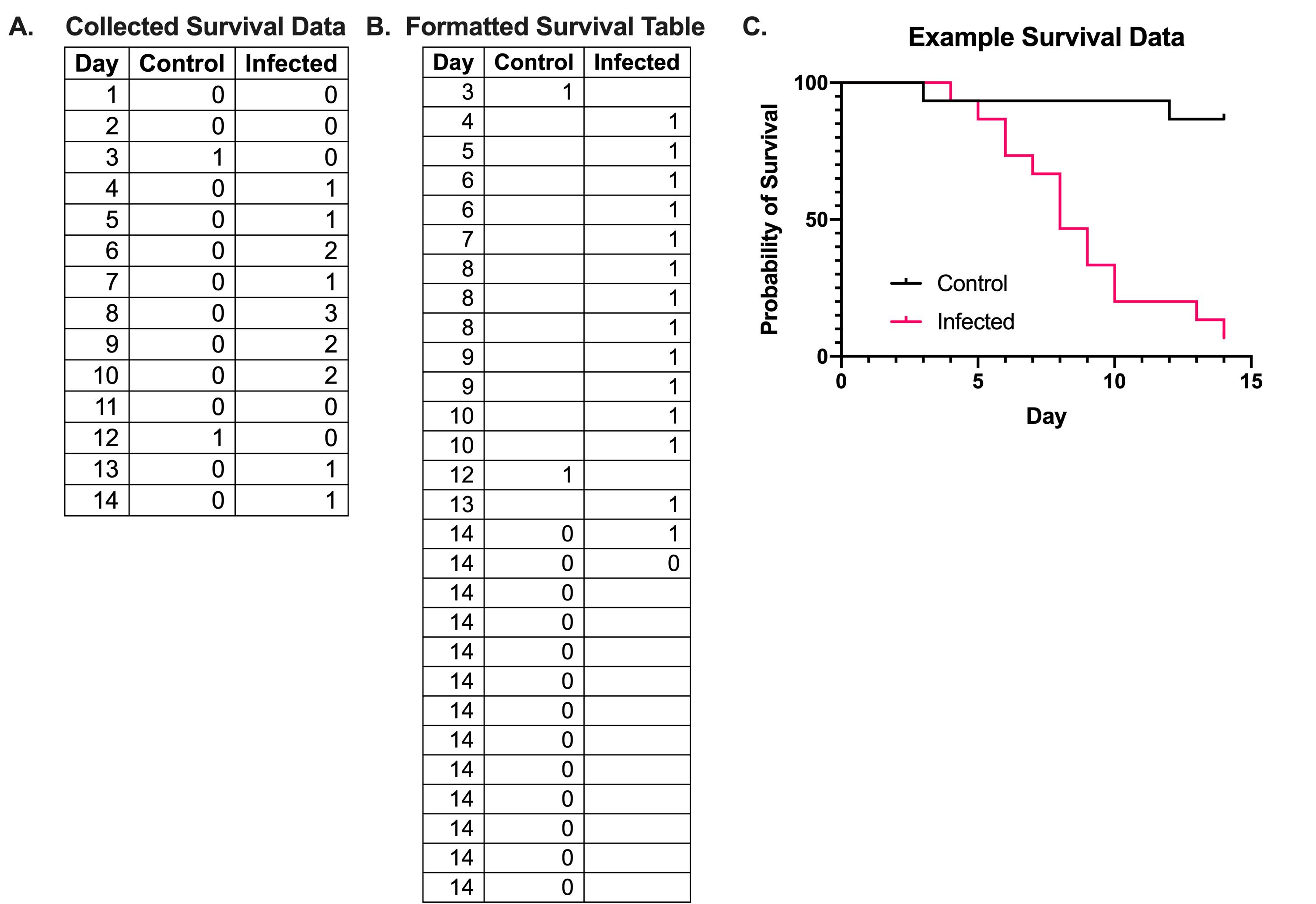
Figure 3. Example formatting and graphing of survival data. (A) Survival data of G. mellonella is collected daily and recorded as the number of deaths that occur in each condition. (B) When converting the survival data into the format required by Prism GraphPad, each event is recorded, with only one event being recorded per line. A death event is recorded as a 1, while a censored event, either due to survival by the end point of the experiment or to the loss of an animal, is marked as a 0. In the control data in (B), there are two death events on days 3 and 12, and 13 censored events on day 14; this indicates that 2 larvae died, while 13 survived until the end of the experiment. The example data from (A, B) is plotted in (C) using Prism GraphPad.
Statistical differences between the tested group can be compared via log rank (Kaplan-Meier) test. P-value of <0.05 indicates a significant difference between datasets. Calculated hazard ratios can also be used to compare virulence between conditions.
Alternative protocol: Virulence assay at 37 °C
Development of cryptococcal infection in G. mellonella larvae can be altered with the changes in surrounding temperature. Incubation of infected larvae at 37 °C helps to mimic human body temperature and reduce the length of the experiment, but simultaneously stimulates metamorphosis of G. mellonella.
Selection of larvae with desirable mass
Select and separate G. mellonella larvae and prepare Petri dishes, one for each tested group. Using a digital scale, select bigger larvae with a body mass between 175 and 300 mg.
Eliminate possibility of contamination by using a sterile tissue soaked with 70% ethanol to clean the proleg area of the larvae before its transfer to the Petri dish.
Preparation of fungal cell inoculum
Approximately 48 h prior to the infection, inoculate 5 mL of YPD media with the desired strain of C. neoformans and incubate on the cell culture rotor at 30 °C.
Pellet the 48 h culture of C. neoformans for 4 min at 2,500 × g in the centrifuge, discard supernatant and provide equal amount (5 mL) of YPD media, and incubate culture for another 2–4 h.
This step allows infections to be performed with the cells in the stage of logarithmic growth, if desired. If not, proceed with the next steps without this additional incubation. Different microbial components, including virulence factors, are expressed at logarithmic stages of growth compared to stationary stages. Performing infections with fungi in logarithmic growth would allow researchers to investigate questions specifically related to how those factors affect virulence.
Transfer 1 mL of refreshed culture to Eppendorf tubes and pellet tested fungal strain by centrifuging tubes for 4 min at 2,500 × g in the tabletop centrifuge.
Wash your samples twice with DPBS, prepare a 1:100 cell dilution, and establish cell concentration using a hemocytometer.
Dilute your washed cells with DPBS to the concentration of 107 cells/mL.
Infection of larvae
Follow the steps of Basic Protocol: Infection of larvae.
Incubation and maintenance of the tested larvae
After finishing series of injections, transfer Petri dishes with larvae to the 37°C incubator and perform evaluation of larvae variability every day by checking the changes in color of each larva or by gently poking the larva using the pointy end of a pipette tip.
Data analysis
Follow the steps of Basic Protocol: Data analysis.
Recipes
Yeast peptone dextrose (YPD) liquid medium
1,000 mL of distilled H2O
20 g BD Bacto Peptone
10 g BD Bacto Yeast Extract
20 g Glucose (Sigma, 1002789701)
Alternatively
1,000 mL dH2O
50 g of Difco YPD Broth
To prepare YPD liquid media, mix all the ingredients in a 2 L Erlenmeyer flask using a magnetic stirring bar. Transfer the mixture into the glass bottles and autoclave for 20 min.
Notes
One main consideration when using Galleria mellonella as a model organism is its biological variability, which will depend on source of larvae or rearing protocols. To date, there is no widely available inbred isogenic G. mellonella strain that serves as a wild type. This opens the model up to genetic variation and thus immunological variability between larvae and larvae sources. Additional significant variability occurs due to diet differences during rearing. For this reason, it is important to perform sufficient replicates with different batches/generations of larvae. When measuring pupal survival, consistency is also important. Prior to emergence as a moth, the pupa will also stop responding to stimuli. This will occur after various amounts of time, depending on the environmental temperature, but will usually occur between days 10–14 at 30°C. While these pupae do not respond to stimulus, they will generally have a lighter color than a dead pupa and a lightweight and soft “dry” feeling when poked. Ideally, the experiment will be completed by this timepoint. Adult moths are censored as data points when they emerge, and survival of moths is not recorded.
Critical Parameters & Troubleshooting
To ensure proper quality of data obtained in the G. mellonella survival assay, it is important to include a negative control group of larvae inoculated with DPBS, as well as a positive control with a well-defined virulent cryptococcal strain (H99 or KN99). High mortality in the negative control group indicates problems with the procedure of injection or low viability of used larvae, which could occur for various reasons. For injection practice, food coloring in water or DPBS could be used to make sure the injections are entering the body cavity, indicated by organism-wide coloration. While there may be researcher-to-researcher variations between defining a G. mellonella larva or pupa as alive or dead, it is important that the threshold and determination of survival are consistent from day-to-day and between replicates. Consistency can be improved by making sure one (or as few as possible) researcher records all survival measurements.
To minimize the risk of accidental needlestick injury, keep a firm grip on the larva, avoid recapping the needle, and dispose all used syringes with needles to the appropriate biohazard sharps containers. In case of a needlestick injury, wash the area, follow procedures established at your research institution, notify the appropriate emergency health clinic at your institution, and seek physician expertise (Casadevall et al., 1994) .
Utilization of G. mellonella as a model organism for investigating C. neoformans virulence allows for a relatively quick assessment of different clinical, environmental, or mutant strains. Results of this assay can often be obtained in less than two weeks. Typically, larvae begin dying rapidly in less than a week following infection with C. neoformans. Increased incubation temperature and initial inoculum of fungal cells can shorten the total time of the experiment.
Acknowledgments
Protocol is adapted from Reeves et al. (2004), Mylonakis et al. (2005), García-Rodas et al. (2011), and Smith et al. (2021). Figures were made using BioRender.com.
Competing interests
We declare no conflict of interest.
Ethics
There are no current ethical restrictions or regulations governing the use of Galleria mellonella in laboratory settings due to their status as invertebrates. G. mellonella were euthanized by freezing for at least 1 h at -20°C.
References
- Almeida, F., Wolf, J. M. and Casadevall, A. (2015). Virulence-Associated Enzymes of Cryptococcus neoformans. Eukaryot Cell 14(12): 1173-1185.
- Browne, N., Heelan, M. and Kavanagh, K. (2013). An analysis of the structural and functional similarities of insect hemocytes and mammalian phagocytes. Virulence 4(7): 597-603.
- Casadevall, A., Mukherjee, J., Yuan, R. and Perfect, J. (1994). Management of injuries caused by Cryptococcus neoformans--contaminated needles. Clin Infect Dis 19(5): 951-953.
- Casadevall, A., Rosas, A. L. and Nosanchuk, J. D. (2000). Melanin and virulence in Cryptococcus neoformans. Curr Opin Microbiol 3(4): 354-358.
- Cogliati, M. (2013). Global Molecular Epidemiology of Cryptococcus neoformans and Cryptococcus gattii: An Atlas of the Molecular Types. Scientifica (Cairo) 2013: 675213.
- Crabtree, J. N., Okagaki, L. H., Wiesner, D. L., Strain, A. K., Nielsen, J. N. and Nielsen, K. (2012). Titan cell production enhances the virulence of Cryptococcus neoformans. Infect Immun 80(11): 3776-3785.
- Eisenman, H. C., Duong, R., Chan, H., Tsue, R. and McClelland, E. E. (2014). Reduced virulence of melanized Cryptococcus neoformans in Galleria mellonella. Virulence 5(5): 611-618.
- Firacative, C., Khan, A., Duan, S., Ferreira-Paim, K., Leemon, D. and Meyer, W. (2020). Rearing and Maintenance of Galleria mellonella and Its Application to Study Fungal Virulence. J Fungi (Basel) 6(3).
- García-Rodas, R., Casadevall, A., Rodriguez-Tudela, J. L., Cuenca-Estrella, M. and Zaragoza, O. (2011). Cryptococcus neoformans capsular enlargement and cellular gigantism during Galleria mellonella infection. PLoS One 6(9): e24485.
- Kwon-Chung, K. J., Fraser, J. A., Doering, T. L., Wang, Z., Janbon, G., Idnurm, A. and Bahn, Y. S. (2014). Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med 4(7): a019760.
- Liu, O. W., Chun, C. D., Chow, E. D., Chen, C., Madhani, H. D. and Noble, S. M. (2008). Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135(1): 174-188.
- Lui, G., Lee, N., Ip, M., Choi, K. W., Tso, Y. K., Lam, E., Chau, S., Lai, R. and Cockram, C. S. (2006). Cryptococcosis in apparently immunocompetent patients. QJM 99(3): 143-151.
- Maurer, E., Hortnagl, C., Lackner, M., Grassle, D., Naschberger, V., Moser, P., Segal, E., Semis, M., Lass-Florl, C. and Binder, U. (2019). Galleria mellonella as a model system to study virulence potential of mucormycetes and evaluation of antifungal treatment. Med Mycol 57(3): 351-362.
- Mylonakis, E., Moreno, R., El Khoury, J. B., Idnurm, A., Heitman, J., Calderwood, S. B., Ausubel, F. M. and Diener, A. (2005). Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 73(7): 3842-3850.
- Pereira, T. C., de Barros, P. P., Fugisaki, L. R. O., Rossoni, R. D., Ribeiro, F. C., de Menezes, R. T., Junqueira, J. C. and Scorzoni, L. (2018). Recent Advances in the Use of Galleria mellonella Model to Study Immune Responses against Human Pathogens. J Fungi (Basel) 4(4).
- Perfect, J. R. and Bicanic, T. (2015). Cryptococcosis diagnosis and treatment: What do we know now. Fungal Genet Biol 78: 49-54.
- Reeves, E. P., Messina, C. G., Doyle, S. and Kavanagh, K. (2004). Correlation between gliotoxin production and virulence of Aspergillus fumigatus in Galleria mellonella. Mycopathologia 158(1): 73-79.
- Smith, D. F. Q. and Casadevall, A. (2021). Fungal immunity and pathogenesis in mammals versus the invertebrate model organism Galleria mellonella. Pathog Dis 79(3).
- Stączek, S., Zdybicka-Barabas, A., Wiater, A., Pleszczynska, M. and Cytrynska, M. (2020). Activation of cellular immune response in insect model host Galleria mellonella by fungal alpha-1,3-glucan. Pathog Dis 78(9).
- Trevijano-Contador, N. and Zaragoza, O. (2018). Immune Response of Galleria mellonella against Human Fungal Pathogens. J Fungi (Basel) 5(1).
- Zaragoza, O. (2019). Basic principles of the virulence of Cryptococcus. Virulence 10(1): 490-501.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Stempinski, P. R., Smith, D. F. Q. and Casadevall, A. (2022). Cryptococcus neoformans Virulence Assay Using a Galleria mellonella Larvae Model System. Bio-protocol 12(15): e4480. DOI: 10.21769/BioProtoc.4480.
Category
Microbiology > Microbe-host interactions > Fungus
Biological Sciences > Biological techniques > Microbiology techniques
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









