- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Auto- and Substrate-Ubiquitination Assays
(*contributed equally to this work) Published: Vol 12, Iss 7, Apr 5, 2022 DOI: 10.21769/BioProtoc.4368 Views: 3452
Reviewed by: Wenrong HeMin CaoYuan Wang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Detection of Disulfides in Protein Extracts of Arabidopsis thaliana Using Monobromobimane (mBB)
Shin‐nosuke Hashida and Maki Kawai-Yamada
Mar 5, 2019 6346 Views
Simple Method to Determine Protein Redox State in Arabidopsis thaliana
Keisuke Yoshida and Toru Hisabori
Jun 5, 2019 7362 Views
Split-luciferase Complementation Imaging Assay to Study Protein-protein Interactions in Nicotiana benthamiana
Liping Wang [...] Rosa Lozano-Durán
Dec 5, 2021 11676 Views
Abstract
The precise regulation of the homeostasis of the cellular proteome is critical for the appropriate growth and development of plants. It also allows the plants to respond to various environmental stresses, by modulating their biochemical and physiological aspects in a timely manner. Ubiquitination of cellular proteins is one of the major protein degradation routes for maintaining cellular protein homeostasis, and ubiquitin E3 ligases, components of ubiquitin ligase complexes, play an important role in the selective degradation of target proteins via substrate-specific interactions. Thus, understanding the role of E3 ligases and their substrate regulation uncovers their specific cellular and physiological functions. Here, we provide protocols for auto- and substrate-ubiquitination analyses that utilize the combination of in vitro purified E3 ubiquitin ligase proteins and immunoprecipitation.
Keywords: UbiquitinationBackground
Ubiquitination is a post-translational modification in which ubiquitins, small regulatory proteins found in most eukaryotes, are covalently attached to cellular proteins, resulting in the proteolysis of the cellular proteins via the 26S proteasomes (Sharma et al., 2016; Cappadocia and Lima, 2018). Ubiquitination consists of sequential and cooperative actions of three enzymes that catalyze the attachment of ubiquitins to substrate proteins: E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin-ligating enzymes. Among these enzymes, E3 ubiquitin-ligating enzymes (also called E3 ubiquitin ligases) provide substrate-specific recognition, which results in the modulation of the homeostasis of a specific cellular protein. The auto- and substrate-ubiquitination are key analyses to understand the function of E3 ubiquitin ligases, as well as their substrate recognition and regulation. Recently, we used auto- and substrate-ubiquitination assays to analyze the functional relationship of Seven-in-Absentia of Arabidopsis 2 (SINAT2), a RING-domain containing E3 ubiquitin ligase involved in stress response, and Arabidopsis ACC synthase 5 (ACS5), a rate-limiting enzyme in ethylene biosynthesis (Lee et al., 2021). Here, we describe protocols for auto-ubiquitination of SINAT2 and SINAT2-mediated ubiquitination of ACS5. Our protocols utilize in vitro purified E3 ubiquitin ligases and immunoprecipitated substrate proteins from a heterogeneously expressed system. Conventionally, substrate ubiquitination assays include purified recombinant substrate proteins and E3 ligases, yielding desired results. However, our protocol utilizes immunoprecipitated substrate proteins expressed from plants. This approach allows researchers to conduct ubiquitination assays for substrate proteins with common problems during recombinant protein expression in E. coli and other expression systems.
Materials and Reagents
Tris-HCl (Dot Scientific Inc., catalog number: DST60040-5000)
CaCl2 (Fisher Bioreagents, catalog number: C79-500)
Adenosine 5’-triphosphate (ATP) (New England Biolabs, catalog number: P0756S)
DTT (Roche, catalog number: 3483-12-3)
MES Free acid monohydrate (Gold Biotechnology, catalog number: M-090-500)
MgCl2 (Fisher Bioreagents, catalog number: BP-214-500)
3',5'-Dimethoxy-4'-hydroxyacetophenone (Acetosyringone) (Acros Organics, catalog number: 115540050)
P19 plasmid
Liquid nitrogen
NaCl (Fisher Bioreagents, catalog number: BP358-10)
EDTA, pH 8.0 (VWR, catalog number: BDH4528-500GP)
Phenylmethaneesulfonyl fluoride (PMSF) (Acros Organics, catalog number: 215740100)
Ammonium persulfate (Acros Organics, catalog number: 327085000)
TEMED (Invitrogen, catalog number: 15524-010)
Glycine (Fisher Bioreagents, catalog number: BP381-5)
SDS micropellets (Fisher Bioreagents, catalog number: BP8200-500)
KCl (Micron Fine Chemicals, catalog number: 6858-04)
Na2HPO4 (Dot Scientific Inc., catalog number: DSS23100-1000)
KH2PO4 (Fisher Bioreagents, catalog number: P285-500)
Bromophenol blue free acid (Gold Biotechnology, catalog number: B-092-5)
2-mercaptoethanol (Acros Brganics, catalog number: 125472500)
Glycerol (Fisher Bioreagents, catalog number: G31-500)
Tween 20 (Fisher Bioreagents, catalog number: BP337-500)
HaltTM Protease inhibitor cocktail, 100× (Thermo Fisher Scientific, catalog number: 78430)
Ubiquitin activating enzyme E1 (Enzo Life Science, catalog number: BML-UW9410-0050)
Ubiquitin conjugating enzyme E2 (Recombinant Human Ubc5b/UBE2D2; R&D Systems, catalog number: E2-622-100)
Recombinant Human Myc-Ubiquitin protein (R&D System, catalog number: U115)
Anti-Ubiquitin (P4D1) antibody (Santa Cruz, catalog number: SC-8017)
Anti-GFP antibody (Roche, catalog number: 1181446001)
Maltose Binding Prtoein polyclonal antibody (Invitrogen, catalog number: PA1-989)
Goat-anti-mouse IgG (H+L) secondary antibody, HRP (Invitrogen, catalog number: 31430)
Goat-anti-rabbit IgG-HRP secondary antibody (Santa Cruz, catalog number: sc-2030)
PierceTM Magnetic A/G agarose beads (Thermo Fisher Scientific, catalog number: 88802)
SDS-PAGE gel (followed by Bio-Rad recipe)
PageRuler prestained protein ladder, 10 to 180 kDa (Thermo Scientific, catalog number: 26617)
Amersham Protran Western blotting membrane sandwich, nitrocellulose (Amersham, catalog number: 19240725)
Phosphate-buffered saline (PBS)
Phosphate-buffered saline with 0.05 % Tween 20 (PBS-T)
Skimmed milk powder (MP Biomedicals, catalog number: 0290288705)
Chemiluminescent HRP substrate kit (Thermo Scientific, catalog number: 34094)
5× SDS sample buffer (see Recipes)
1×SDS running buffer (see Recipes)
1× Transfer buffer (see Recipes)
1× Phospated Buffered Saline (PBS-T with 0.05% Tween-20) (see Recipes)
3% blocking buffer (see Recipes)
Tobacco infiltration buffer (see Recipes)
Immunoprecipitation buffer (see Recipes)
Equipment
Refrigerated centrifuge (Labnet, model: C2500-R)
Sonicator (Fisher brand, model: F1350)
Protein electrophoresis apparatus (Bio-Rad, Mini-PROTEAN tetra handcast system and PowerPac Poweer Supplies)
Protein transfer system (Trans-Blot Turbo System, model: 1704150)
Benchtop rocking shaker (Labnet, model: orbit1000)
Benchtop tube rotator (Fisher brand, model: 88861051)
Procedure
Auto-ubiquitination analysis of MBP-SINAT2
Prepare ubiquitination reaction mixtures in a total volume of 30 μL, consisting of 40 mM Tris-HCl, pH 7.5, 5 mM CaCl2, 2 mM ATP, 2 mM DTT, 50 ng of E1, 250 ng of E2, 1 µg of ubiquitin, and 500 ng of purified recombinant protein (MBP, MBP-SINAT2, or MBP-SINAT2C63S). Set up two ubiquitination control reactions for Maltose-Binding Protein (MBP), MBP-SINAT2, or MBP-SINAT2C63S, by adding the ubiquitination reaction mixture without E1 or E2 (Figure 1).
Note: MBP-SINAT2C63S is an inactive form of SINAT2. The substitution of Cys53 for Ser results in abolishing the E3 ligase activity of SINAT2.
Incubate the mixtures at 30°C with agitation for 1 h, by placing Eppendorf tubes containing the reaction mixture on a benchtop tube rotator.
Add 5× SDS sample buffer to the mixtures to stop the reaction.
Apply the reactions and the prestained protein marker (2.5 µL) to an 8% SDS-PAGE gel. Separate the reaction samples by electrophoresis in 1× SDS running buffer at 120 V for 150 min.
Transfer the proteins in the SDS-PAGE gel to a nitrocellulose membrane in 1× transfer buffer, using the Bio-Rad Transblot Turbo transfer system at room temperature and 25 V for 30 min.
Remove the membrane from the transfer system and place it in a plastic container (7 cm × 9 cm). Briefly, wash the membrane with 1× PBS-T.
Block the membrane with 3% blocking buffer at room temperature with gentle agitation using a rocking shaker for 1 h, and subsequently add the primary anti-MBP or anti-ubiquitin antibody to the blocking buffer, in the following dilutions: 1:10,000 for the anti-MBP, and 1:3,000 for the anti-ubiquitin antibody. Incubate the membrane with the primary antibody solution with gentle agitation using a rocker at room temperature for 1 h.
Rinse the membrane with 1× PBS-T with gentle agitation on a rocker at room temperature four times for 10 min each.
To detect anti-MBP and anti-ubiquitin, place the membrane in a plastic container containing a 3% blocking buffer. Add a dilution of 1:5,000 of secondary antibody, and incubate the membrane with gentle agitation on a rocker at room temperature for 50 min.
Rinse the membrane with 1× PBS-T with gentle agitation on a rocker at room temperature four times for 10 min each.
Place the membrane on plastic wrap, evenly apply the chemiluminescence solution, and incubate for 3 min.
Remove the excess chemiluminescence solution from the membrane, wrap the membrane with plastic wrap, and place it in an X-ray cassette.
Add an X-ray film to the a cassette, and develop the film using a film processor.
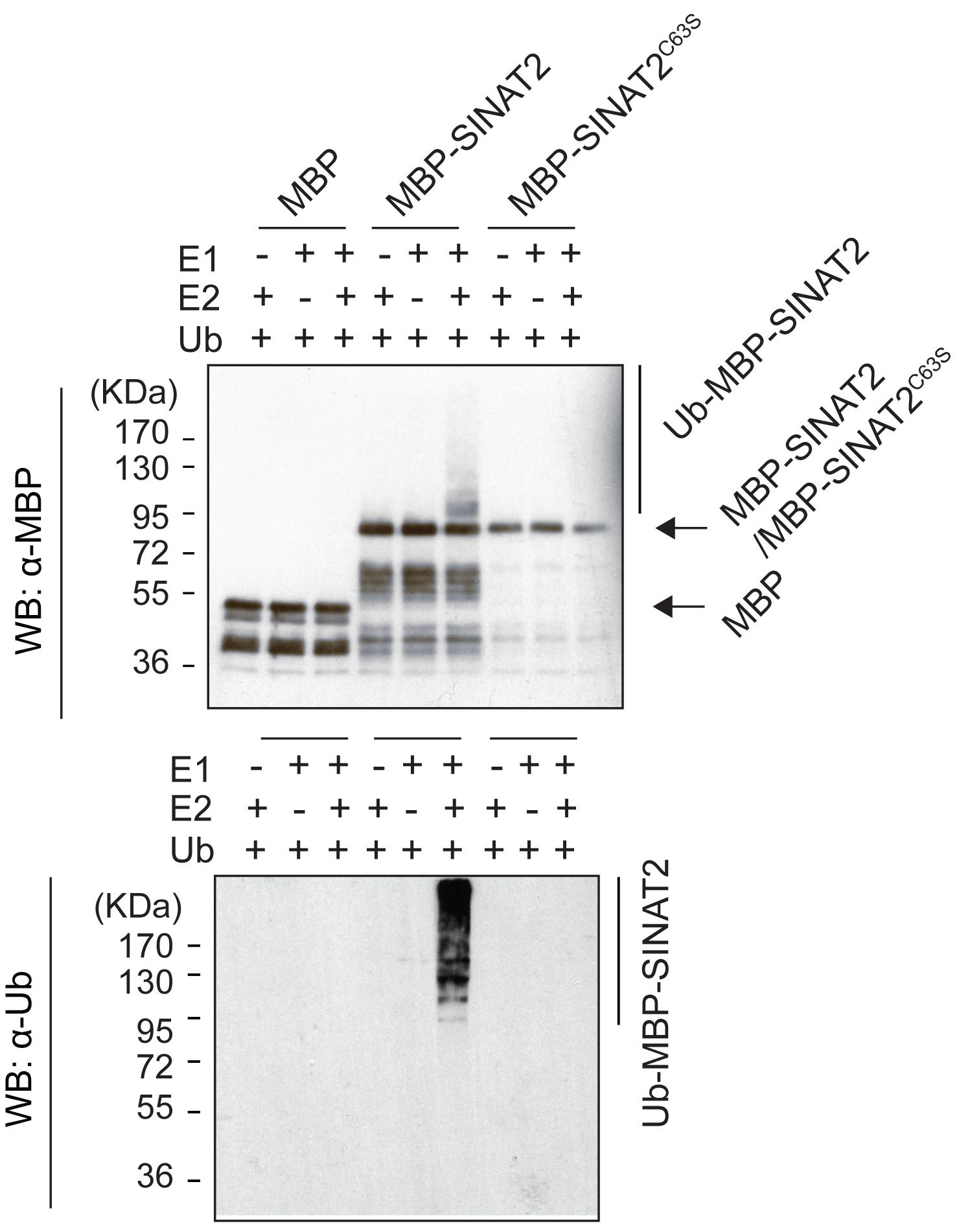
Figure 1. Auto-ubiquitination of SINAT2. MBP-SINAT2 E3 ligase was auto-ubiquitinated in the presence of E1, E2, and Ubiquitin proteins, whereas free MBP and inactive MBP-SINAT2C63S failed for autoubiquitination (modified from Lee et al., 2021).
Substrate-ubiquitination analysis of immunoprecipitated substrate proteins
Infiltrate tobacco (Nicotiana benthamiana) grown in a growth chamber (16-h-light/8-h-dark cycle; 28°C/28°C) with Agrobacterium tumefaciens transformed with YFP-ACS5 plasmid DNA and P19, and incubate them at 28°C for 3 days.
Excise a portion of the infiltrated tobacco leaves with a laser blade and observe the YFP fluorescence of YFP-ACS5, using an epifluorescence microscope. If the YFP fluorescence is widely observed throughout the infiltrated leaves, proceed to step 3; otherwise, repeat step 1 until YFP fluorescence is observed (Figure 2).
Extract the total protein from two infiltrated tobacco leaves, by grinding the leaves using liquid nitrogen followed by homogenizing the grounded tissue powder in 300 μL of coimmunoprecipitation buffer (see Recipes).
Centrifuge at 12,400 × g and 4°C for 15 min, and transfer 200 μL of the supernatant containing the total protein extract to a new microcentrifuge tube.
The substrate ubiquitination reaction is performed in a total volume of 50 μL, consisting of 40 mM Tris-HCl, pH 7.5, 5 mM CaCl2, 2 mM ATP, 2 mM DTT, 50 ng of E1, 250 ng of E2, 1 μg of ubiquitin, a total of 40 μL of the protein extract from step 4 containing the YFP-ACS5 protein, and 500 ng of purified MBP-SINAT2 (or MBP, or MBP-SINAT2C63S). Free MBP and inactive MBP-SINAT2C63S serve as negative controls. Aliquot 10 µL of each reaction for input analysis (Figure 3).
Incubate the mixture with gentle rotation using a benchtop lab rotator at 30°C for 1 h.
Add 5 µg of anti-GFP and protein A/G agarose magnetic beads (20 µL) into the ubiquitination reaction tubes from step 6, and incubate with gentle agitation at 4°C overnight.
Collect the protein A/G magnetic beads using a magnetic stand, and wash the reaction mixture three times with immunoprecipitation buffer (see Recipes).
Elute the bound proteins from protein A/G agarose magnetic beads with 2× SDS sample buffer.
Perform electrophoresis and western blotting as described in the auto-ubiquitination analysis of MBP-SINAT2 (steps 4 to 13), and detect ubiquitinated YFP-ACS5 and input proteins using anti-MBP, anti-ubiquitin, and anti-GFP antibodies.
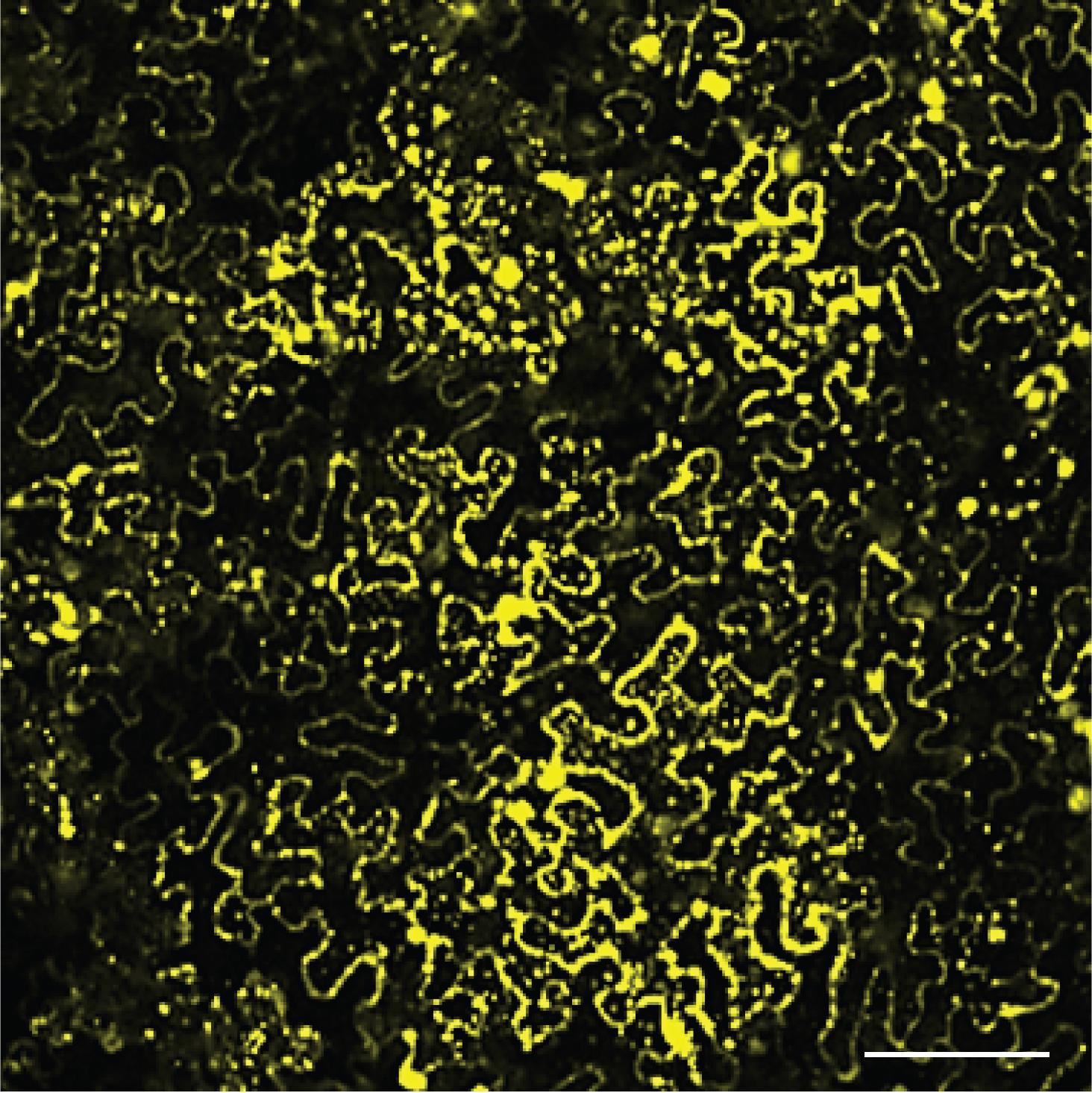
Figure 2. Expression of YFP-ACS5 in N. benthamiana leaves. N. benthamiana leaves were infiltrated with Agrobacteria transformed with a YFP-ACS5 plasmid and incubated for 3 days. The fluorescence of YFP-ACS5 was widely observed throughout the leaf by confocal microscopy. Scale bar, 50 µm.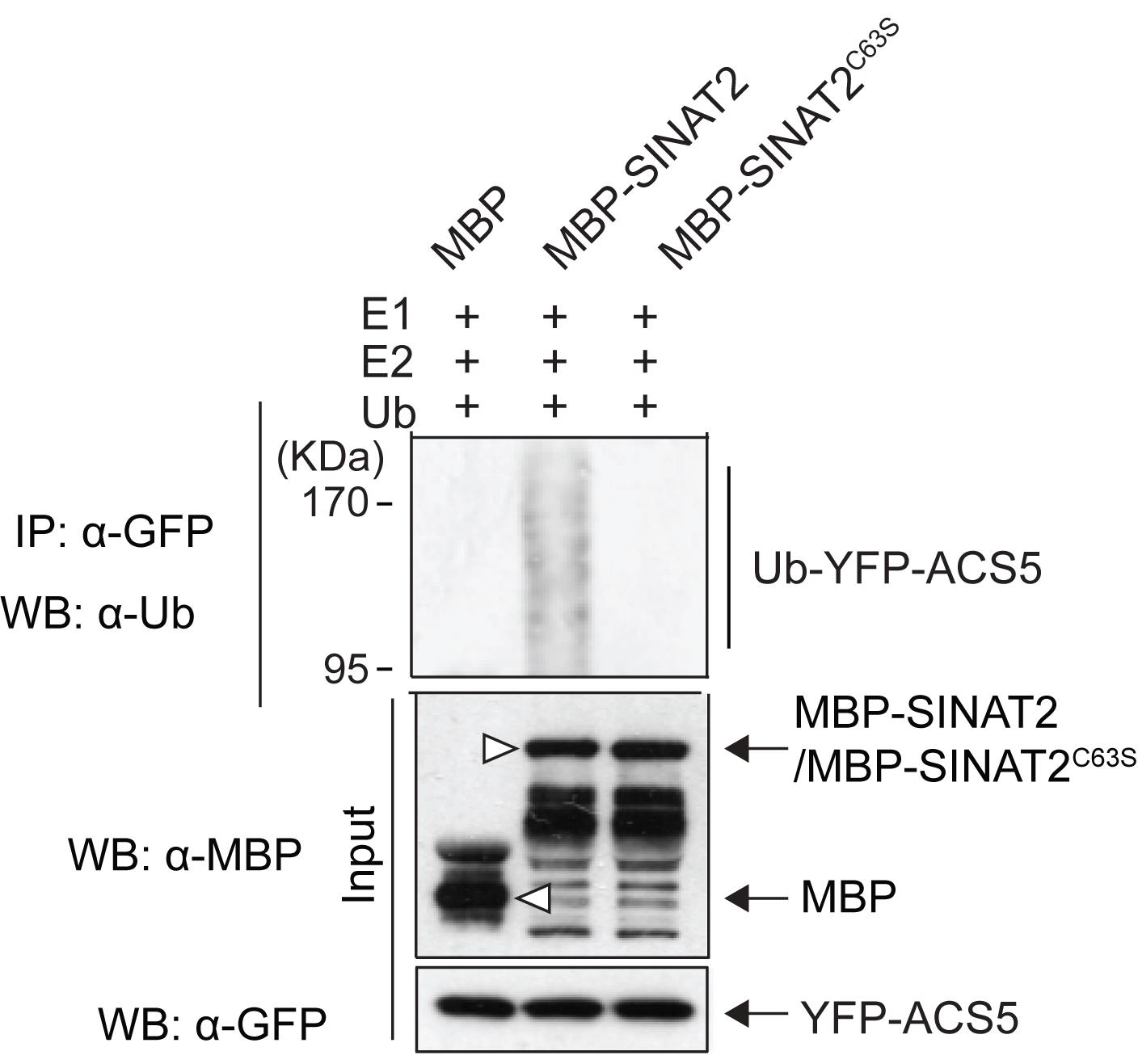
Figure 3. SINAT2 E3 ligase-mediated substrate ubiquitination of ACS5. The ubiquitination of YFP-ACS5 is only detected in the presence of active MBP-SINAT2 E3 ligase proteins. Free MBP and MBP-SINAT2C63S, an inactive form of SINAT2, are negative controls. Open triangles indicate free MBP and MBP-fused SINAT2 proteins (modified from Lee et al., 2021).
Recipes
5× SDS sample buffer
10% SDS
30% glycerol
0.02% bromophenol blue
250 mM Tris-Cl, pH 6.8
5% 2-mercaptoethanol
1× SDS running buffer
25 mM Tris
192 mM Glycine
0.1% SDS
pH 8.3
1× Transfer buffer
25 mM Tris
192 mM Glycine
20 % methanol (V/V)
0.05 % SDS
1× Phospated Buffered Saline (PBS-T with 0.05% Tween-20)
137 mM NaCl
2.7 mM KCl
10 mM Na2HPO4
1.8 mM KH2PO4
3% blocking buffer
3% skimmed milk powder (w/v) in PBS-T
Tobacco infiltration buffer
10 mM MES, pH 5.6
10 mM MgCl2
100 µM Acetosyringone
Immunoprecipitation buffer
25 mM Tris, pH 7.5
150 mM NaCl
5 mM EDTA, pH 8.0
1 mM DTT
1 mM PMSF
1× protease inhibitor cocktail
Acknowledgments
This work was supported by the Purdue University start-up fund and NSF (MCB 1817286) to GMY. This protocol was adapted from the research article of Lee et al. (2021).
Competing interests
The authors declare no competing interest for this study.
References
- Cappadocia, L. and Lima, C. D. (2018). Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism. Chem Rev 118(3): 889-918.
- Lee, H. Y., Park, H. L., Park, C., Chen, Y. C. and Yoon, G. M. (2021). Reciprocal antagonistic regulation of E3 ligases controls ACC synthase stability and responses to stress. Proc Natl Acad Sci U S A 118(34): e2011900118.
- Sharma, B., Joshi, D., Yadav, P. K., Gupta, A. K. and Bhatt, T. K. (2016). Role of Ubiquitin-Mediated Degradation System in Plant Biology. Front Plant Sci 7: 806.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Park, H. L., Lee, H. Y. and Yoon, G. M. (2022). In vitro Auto- and Substrate-Ubiquitination Assays. Bio-protocol 12(7): e4368. DOI: 10.21769/BioProtoc.4368.
Category
Plant Science > Plant biochemistry > Protein > Modification
Biochemistry > Protein > Activity
Biochemistry > Protein > Interaction > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










