- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fractionation and Extraction of Crude Nuclear Proteins From Arabidopsis Seedlings
Published: Vol 12, Iss 2, Jan 20, 2022 DOI: 10.21769/BioProtoc.4296 Views: 6619
Reviewed by: Zhibing LaiKumiko OkazakiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimized Isolation of Lysosome-Related Organelles from Stationary Phase and Iron-Overloaded Chlamydomonas reinhardtii Cells
Jiling Li and Huan Long
Nov 20, 2024 1766 Views

Isolation and Biophysical Characterization of Extracellular Vesicles From Hairy Root Cultures
Marisa Conte [...] Alfredo Ambrosone
Mar 5, 2025 2218 Views

Rapid Miniprep of Intact Chloroplasts from Arabidopsis thaliana Leaves
Brenda A. Carranza-Correa [...] Manuel Gutiérrez-Aguilar
May 20, 2025 2645 Views
Abstract
The plant nucleus is an important subcellular organelle that contains the genome, ribosomal RNA, and regulatory proteins, and performs a central role in the functioning and metabolism of the cell. Fractionation of intact nuclei is a crucial process to elucidate the function of nuclear proteins. Here, we present a simple method for the fractionation of crude nuclei and extraction of nuclear proteins, based on previously established methods. This protocol provides an easy and quick method to isolate crude nuclei and extract nuclear proteins from Arabidopsis seedlings, which is useful for the research on the nuclear proteins, without requirement for high-purity nuclei.
Graphic abstract:

Schematic procedure for the isolation of crude nuclei and extraction of nuclear proteins from Arabidopsis seedlings.
Background
The plant nucleus is the repository of a cell that regulates the genetic and biochemical machinery, therefore it is crucial to isolate the plant nuclei as source material for proteomic studies, protein expression patterns, modifications, and dynamic physiological responses (Calikowski and Meier, 2006). Nuclei isolation from different plant tissues mainly involves four steps, namely disruption, filtration, repeated centrifugation, and density gradient separation (Yin and Komatsu, 2016). Homogenization buffers are important for the stabilization of nuclei from various plant tissues. The compositions of homogenization buffers includes organic buffers (e.g., MES, HEPES, PIPES, and Tris) to stabilize the pH, inorganic salts (e.g., KCl, NaCl, and MgCl2) to maintain the ionic strength of the solution, sucrose, glycerol, and hexylene glycol to stabilize membranes, polyamines (spermine and spermidine) to stabilize the nuclear chromatin and avoid the aggregation of nuclei, reducing agents (β-mercaptoethanol and dithiothreitol) to maintain cysteine residues in the reduced form, and protease inhibitors (phenylmethylsulfonyl fluoride and plant protease inhibitor cocktail) to protect proteins from degradation (Yin and Komatsu, 2016). For the fact that various organelles of a cell differ in both size and density, density gradient centrifugation (also known as rate-zonal centrifugation) and differential-velocity centrifugation are widely used for the purification of different subcellular organelles (Komatsu, 2007). Percoll density gradient is often used in the fractionation and purification of nuclei from cell homogenates of Arabidopsis seedlings (Calikowski and Meier, 2006). The 2.0 M sucrose gradient method is also rapid and efficient for the isolation of nuclei from the cultured rice suspension cells (Morré and Andersson, 1994). Here, we present a simple protocol for the fractionation of the crude nuclei from Arabidopsis seedlings without density gradient centrifugation, based on the previously established methods from Calikowski and Meier (2006) and Liu et al. (2018). This method is suitable for analyzing accumulation, as well as nuclear and cytoplasmic distributions of specific proteins under specific stress conditions (Wang et al., 2011; Liu et al., 2018; Xu et al., 2021). However, percoll density gradient centrifugation is recommended, if high-quality nuclear proteins are needed.
Materials and Reagents
Materials
200 μL pipette tips
1 mL pipette tips
5 mL pipette tips
100-μm Nylon cell stainer (Corning Incorporated, FALCON, catalog number: 352350)
40-μm Nylon cell stainer (Corning Incorporated, FALCON, catalog number: 352340)
50 mL centrifuge tube
1.5 mL centrifuge tube
Arabidopsis seedlings
Liquid nitrogen
Reagents
Sucrose (Sinopharm, SCR, catalog number: 10021418)
Ficoll 400 (Sigma-Aldrich, catalog number: F9378-5G)
Dextran T40 (Solarbio, catalog number: D8250)
Tris (Sangon Biotech, catalog number: A610195)
HCl (Sinopharm, SCR, catalog number: 10011018)
MgCl2 (RHAWN, catalog number: R007504-500 g)
Triton X-100 (Sigma-Aldrich, catalog number: V900502-100ML)
phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: PMSF-RO)
β-mercaptoethanol (Sigma-Aldrich, catalog number: 07604-100ML)
Protease inhibitor cocktail (Roche, cOmplete Tablets EDTA-free, EASYpack, catalog number: 04693132001)
KCl (Sinopharm, SCR, catalog number: 10016318)
HEPES (Sigma-Aldrich, catalog number: H3375)
Hexylene glycol (Sangon Biotech, catalog number: A607060)
Spermine (Sigma-Aldrich, catalog number: S3256)
Spermidine (Sigma-Aldrich, catalog number: S2626)
Glycerol (Sangon Biotech, catalog number: A100854)
NaCl (Sinopharm, SCR, catalog number: 10019318)
0.5 M EDTA, pH 8.0 (Biosharp, catalog number: 69075458)
Dithiothreitol (DTT) (Beyotime, catalog number: ST043-5g)
Urea (Sangon Biotech, catalog number: A600148)
Nonidet P-40 (NP-40) (Roche, catalog number: 11332473001)
Anti-Histone H3 antibody (Abcam, catalog number: ab18521)
Stock solution
Stock solutions for the buffers can be made before and stored at 4°C (1-5) or -20°C (6-10). On the day of the experiment, the buffers can be diluted as required.
1 M Tris-HCl, pH 7.4
20% Triton X-100 (v/v)
20% NP-40 (v/v)
2 M KCl
0.5 M HEPES-KOH, pH 7.0
50 mM Spermine
125 mM Spermidine
10 mM PMSF
1.0 M DTT
Protease inhibitor cocktail (100×) (see Recipes)
Honda buffer (see Recipes)
Nuclei isolation buffer (NIB) buffer (see Recipes)
Glycerol buffer (see Recipes)
Nuclei lysis buffer (see Recipes)
Equipment
Centrifuge (ThermoFisher, catalog number: 75004380)
Vortex shaker
Mortar
Procedure
Nuclei isolation
Transfer 2.0 g of the 10-day-old Arabidopsis seedlings into a mortar, which is prechilled with liquid nitrogen. Grind in liquid nitrogen for about 5 min until a fine powder is obtained. Transfer the frozen seedling powder into a 50 mL tube filled with 4 mL of precooled Honda buffer (2 mL/g), mix by vortex, then incubate on ice for 10 min.
Filter the homogenate through a 100-μm and then a 40-μm mesh nylon cell stainer sequentially, take 100 μL of the flow-through as the total protein extract (control 1), then centrifuge the remaining flow-through at 1,500 × g for 5 min at 4°C.
Transfer 100 μL of the supernatant to a new tube, then centrifuge at 12,000 × g for 10 min at 4°C, collect the supernatant as the cytosol fraction (control 2).
Discard the remaining supernatant, wash the dark-green pellet containing a crude nuclear fraction with 5 mL of nuclei isolation buffer (NIB). Gently pipet up and down with a 5 mL-sterile pipette. Centrifuge at 1,500 × g for 10 min at 4°C, discard the supernatant, and pellet the nuclei.
Repeat Step A4 three or four times, until the color of the nuclear pellet becomes white, with as little green coloration as possible. The white nuclear pellet is regard as the crude nuclei fraction.
Nuclear protein extraction
Gently resuspend the nuclear pellet in 30 μL of prechilled glycerol buffer and 30 μL of nuclei lysis buffer, then mix the nuclei pellet by vortexing, and incubate on ice for 10 min, to obtain the lysis of the nucleus as the nuclear fraction.
Add 12 μL of 6× SDS protein loading buffer and mix with the nucleus fraction by vortexing, then boil at 100°C for 10 min. Centrifuge at 12,000 × g for 10 min at room temperature. Aspirate the supernatant (nuclear proteins) to a new tube, store the sample at -80°C untill use.
Data analysis
Before nuclear protein extraction, check the quality of nuclei through 4',6-diamidino-2-phenylindole (DAPI) (0.33 μg/mL) staining of the crude nuclei, and estimate the ratio of intact nuclei under a fluorescent microscope immediately after staining (Figure 1A).
Estimate the quality of nuclear proteins by western blot using anti-Histone H3 antibody (Abcam, USA, catalog number: ab18521), using the total protein extract (control 1) and the cytosol fraction (control 2) as controls. Histone H3 is found in the total protein and nucleus fractions, but not in the cytosol fraction, suggesting that the nucleus fraction is enriched in nuclear proteins (Figure 1B).
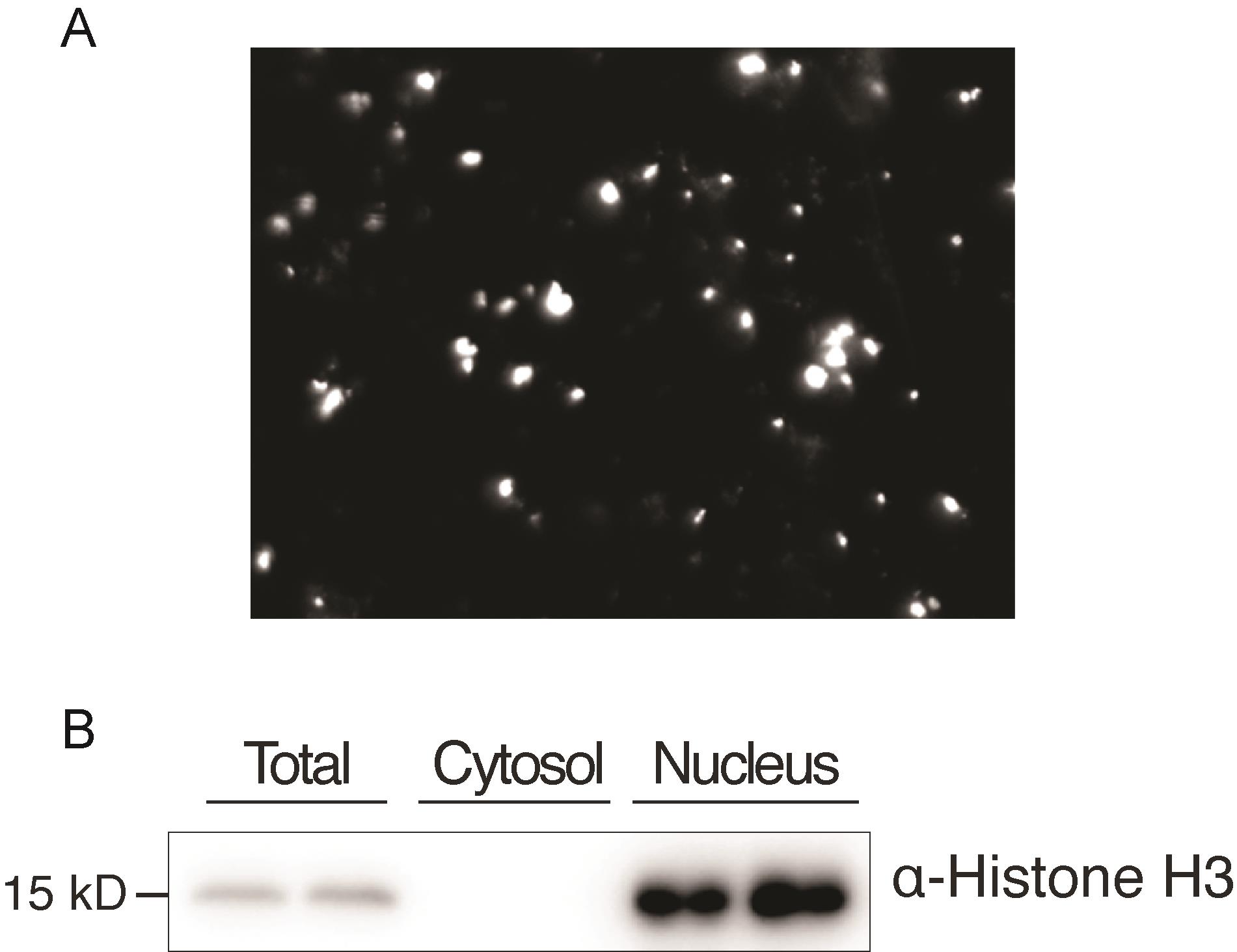
Figure 1. Estimation of the quality of nuclear proteins. (A) Observation of nuclei (white points) under a fluorescent microscope after DAPI staining. (B) Western blot analysis using anti-Histone H3 antibody. The total protein extract (total), the nuclear fraction (nucleus), and the fraction devoid of nucleus (cytosol).
Notes
All the steps for isolation of nuclei are performed on ice or at 4°C.
Add protease inhibitors (e.g., PMSF and protease inhibitor cocktail) to extraction solutions at the moment before you start.
Recipes
Protease inhibitor cocktail (100×)
Dissolve one tablet in 500 μL of ddH2O.
Honda buffer
0.4 M Sucrose
2.5% Ficoll
5% Dextran T40
25 mM Tris-HCl, pH 7.4
10 mM MgCl2
0.5% Triton X-100
0.5 mM phenylmethylsulfonyl fluoride (PMSF)
10 mM β-mercaptoethanol
1× Plant protease inhibitor cocktail
Nuclei isolation buffer (NIB) buffer
20 mM KCl
20 mM HEPES, pH 7.4
0.5% Triton X-100
13.8% hexylene glycol
0.1% (v/v) β-mercaptoethanol
50 μM spermine
125 μM spermidine
1 mM PMSF
1× Plant protease inhibitor cocktail
Glycerol buffer
20 mM Tris-HCl, pH 7.9
50% (v/v) glycerol
75 mM NaCl
0.5 mM EDTA
0.85 mM Dithiothreitol (DTT)
0.125 mM PMSF
1× Plant protease inhibitor cocktail
Nuclei lysis buffer
10 mM HEPES, pH 7.6
1 mM DTT
7.5 mM MgCl2
0.2 mM EDTA
0.3 M NaCl
1 M Urea
1% Nonidet P-40 (NP-40)
0.5 mM PMSF
1× Plant protease inhibitor cocktail
Acknowledgments
This protocol was adapted from the methods described in Liu et al. (2018) and Calikowski and Meier (2006).
Competing interests
The authors declare no conflicts of interest.
References
- Calikowski, T. T. and Meier, I. (2006). Isolation of Nuclear Proteins. In: Arabidopsis Protocols. Salinas, J. and Sanchez-Serrano, J. J. (Eds.). Humana Press: Totowa, NJ. pp. 393-402.
- Komatsu, S. (2007). Extraction of Nuclear Proteins. In: Plant Proteomics: Methods and Protocols. Thiellement, H., Zivy, M., Damerval, C., and Méchin, V. (Eds.). Methods in Molecular Biology. Humana Press: Totowa, NJ. pp. 73-77.
- Liu, C., Xin, Y., Xu, L., Cai, Z., Xue, Y., Liu, Y., Xie, D., Liu, Y. and Qi, Y. (2018). Arabidopsis ARGONAUTE 1 Binds Chromatin to Promote Gene Transcription in Response to Hormones and Stresses. Dev Cell 44(3): 348-361 e347.
- Morré, D. J. and Andersson, B. (1994). Isolation of all major organelles and membranous cell components from a single homogenate of green leaves. Meth Enzymol 228(6): 412-419.
- Wang, W., Ye, R., Xin, Y., Fang, X., Li, C., Shi, H., Zhou, X., and Qi, Y. (2011). An Importin β Protein Negatively Regulates MicroRNA Activity in Arabidopsis. Plant Cell 23: 3565-3576.
- Xu, F., Jia, M., Li, X., Tang, Y., Jiang, K., Bao, J., and Gu, Y. (2021). Exportin-4 coordinates nuclear shuttling of TOPLESS family transcription corepressors to regulate plant immunity. Plant Cell 33: 697-713.
- Yin, X. and Komatsu, S. (2016). Plant nuclear proteomics for unraveling physiological function. N Biotechnol 33(5 Pt B): 644-654.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhao, J., Bao, J. and Xu, F. (2022). Fractionation and Extraction of Crude Nuclear Proteins From Arabidopsis Seedlings . Bio-protocol 12(2): e4296. DOI: 10.21769/BioProtoc.4296.
Category
Plant Science > Plant cell biology > Organelle isolation
Cell Biology > Organelle isolation > Nuclei
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link