- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Optimised Method for the Production and Titration of Lentiviral Vectors Pseudotyped with the SARS-CoV-2 Spike
Published: Vol 11, Iss 16, Aug 20, 2021 DOI: 10.21769/BioProtoc.4194 Views: 4747
Reviewed by: George William CarnellRan ChenShyam Solanki

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3000 Views

Artificial Metalloenzymes in Artificial Sanctuaries Through Liquid–Liquid Phase Separation
Kaixin Wang [...] Tong Wu
Oct 5, 2025 1536 Views
FLARE: A Flow Cytometry–Based Fluorescent Assay for Measuring HSV-1 Nuclear Egress
Bing Dai [...] Ekaterina E. Heldwein
Jan 5, 2026 512 Views
Abstract
The use of recombinant lentivirus pseudotyped with the coronavirus Spike protein of SARS-CoV-2 would circumvent the requirement of biosafety-level 3 (BSL-3) containment facilities for the handling of SARS-CoV-2 viruses. Herein, we describe a fast and reliable protocol for the transient production of lentiviruses pseudotyped with SARS-CoV-2 Spike (CoV-2 S) proteins and green fluorescent protein (GFP) reporters. The virus titer is determined by the GFP reporter (fluorescent) expression with a flow cytometer. High titers (>1.00 E+06 infectious units/ml) are produced using codon-optimized CoV-2 S, harbouring the prevalent D614G mutation and lacking its ER retention signal. Enhanced and consistent cell entry is achieved by using permissive HEK293T/17 cells that were genetically engineered to stably express the SARS-CoV-2 human receptor ACE2 along with the cell surface protease TMPRSS2 required for efficient fusion. For the widespread use of this protocol, its reagents have been made publicly available.
Graphic abstract:
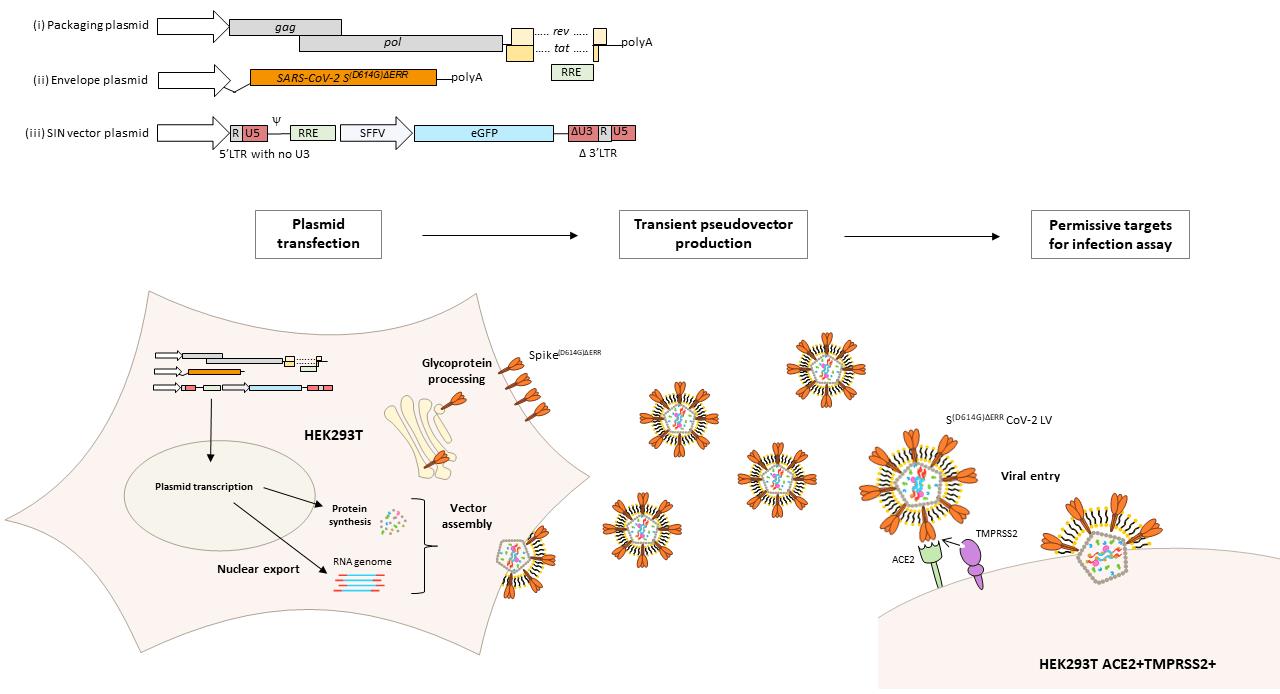
Production and quantification of lentiviral vectors pseudotyped with the SARS-CoV-2 Spike glycoprotein
Background
Studies on SARS-CoV-2 viruses are hampered by the difficulty to produce and manipulate the live viruses that require biosafety level 3 (BSL-3) labs. An alternative to using live virus is to use recombinant lentivirus pseudotyped with the SARS-CoV-2 Spike protein. Pseudotyped viral vectors are very powerful tools for studying biological processes related to enveloped viruses, such as viral entry and immunological response. Pseudotyped viral particles consist of the envelope glycoprotein of one virus with a replication-deficient core of another virus. This allows the deficient core to be dependent on the pseudotyping envelope for target cell entry, thus allowing the investigation of SARS-CoV-2 infection and related serological responses.
Spike is a type-I fusion transmembrane protein expressed on the surface of viral particles as a crown-shaped trimer of heterodimers. In host cells, precursor glycoproteins are proteolytically cleaved by furin at the multibasic S1/S2 site, resulting in dimers composed of an extracellular subunit (S1) containing the receptor-binding domain, which is non-covalently attached to a transmembrane subunit (S2) responsible for viral fusion and subsequent cell entry (Hoffmann et al., 2020a; Peacock et al., 2020). Spike is both necessary and sufficient to induce membrane fusion and cell entry by first binding to its human receptor, ACE2 (hACE2), followed by its proteolytic cleavage by target cell proteases such as the transmembrane protease serine 2 (TMPRSS2) (Hoffmann et al., 2020b; Walls et al., 2020). Thus, Spike has been shown to be the primary target for neutralizing antibodies in COVID-19 convalescent patient sera (Chen et al., 2020; Ju et al., 2020; Pinto et al., 2020).
Spike from different coronaviruses have been successfully pseudotyped on different non-replication competent viruses (Carnell et al., 2017; Grehan et al., 2015; Yan et al., 2007; Fukushi et al., 2005). Accordingly, different viral vectors have been pseudotyped with Spike from SARS-CoV-2, including VSV (Letko et al., 2020; Nie et al., 2020), HIV (Ou et al., 2020; Wu, 2020), and MLV (Pinto et al., 2020; Quinlan et al., 2020) based vectors for neutralization assays, which have been reported to correlate with the live strain. However, a detailed protocol of pseudovector production has not been described for widespread application.
Herein, we describe a fast and reliable protocol for the production of a self-inactivating lentiviral vector pseudotyped with SARS-CoV-2’s Spike glycoprotein and expressing enhanced green fluorescent protein (eGFP) as a marker of infection, which has recently been used to determine neutralisation efficiency of COVID-19 therapeutics on four SARS-CoV-2 variants (Ferrari et al., 2021). The protocol employs a three-plasmid transfection in HEK293T/17 cells with the following plasmids: (i) plasmid expressing the HIV-1 lentiviral genes gag, pol, rev, and tat: (ii) a self-inactivating transfer vector encoding eGFP under an internal viral promoter derived from the spleen focus-forming virus (SFFV); and (iii) a plasmid encoding codon-optimized Spike, with or without the prevalent D614G mutation (Korber et al., 2020), under the cytomegalovirus (CMV) promoter with its ER retention signal (amino acid residues 1255 to 1273) deleted as it has been shown to enhance surface expression (Ou et al., 2020). Produced vectors are then quantified by an infectivity assay on genetically engineered HEK293T/17 cells that stably express hACE2 and TMPRSS2 (Supplementary Figure 1). This protocol has been recently used to produce lentiviral vectors pseudotyped with different SARS-CoV-2's Spike variants. These cells have been deposited in the repository of the National Institute for Biological Standards and Control’s Covid-19-related research reagents (CFAR #101008) for widespread use.
Materials and Reagents
Filtered Pipette tips 1-10, 1-20, 1-200, 100-1,000 μl (Starlab, catalog numbers: S1120-3810 [P10]; S1120-1810 [P20]; S1120-8810 [P200]; S1122-1830 [P1000]) or equivalent
10 cm plates (tissue-culture treated 100 mm dish; Corning, catalog number: 430167) or equivalent
24-well plates (TC-treated 24-well plate; Corning® Costar®, catalog number: 3527) or equivalent
Microcentrifuge tube with screw caps write-on graduated with lid latch 1.5 ml
15 ml conical centrifuge tubes (Merck, Corning®, catalog number: CLS430791) or equivalent
NuncTM EasYFlaskTM 75 cm2 cell culture flasks (ThermoFisher, catalog number: 156472) or equivalent
Sterile syringes (20 ml), generic
Millex-HV syringe filter unit, 0.45 µm, PVDF, 33 mm, gamma sterilized (Merck, catalog number: SLHV033RB) or equivalent
96-well V-bottom plate (96 Well TC-treated microplates, Corning, catalog number: 3894) or equivalent
Dry ice
Ethanol absolute ≥99.8% (Avantor, VWR Chemicals, catalog number: 20821.467DP) or equivalent
HEK 293T/17 cells (ATCC, catalog number: CRL-11268).
Engineered HEK 293T/17 cells with human ACE2 and TMPRSS2 (NIBSC code CFAR#101008)
Plasmids:
Transfer vector: pCCL.SFFV.eGFP
Lentiviral transfer vector encoding eGFP downstream of an SFFV promoter in a pCCL backbone.
Second-generation lentiviral packaging plasmid: pCMVR8.74 (Addgene, catalog number: 22036)
Glycoprotein expression plasmid: pCDNA-SARS-CoV-2-S(D614G)ΔERR
SARS-CoV-2 Spike glycoprotein downstream a CMV promoter in a pCDNA backbone. The glycoprotein contains the D614G substitution and deletion of the last 19 amino acids on its carboxyl-terminus to remove its ER retention signal (ΔERR).
Iscove’s Modified Dulbecco’s Medium (Merck, Sigma-Aldrich, catalog number: I3390) supplemented with 2mM L-glutamine and 10% fetal bovine serum (FBS) (Biosera, catalog number: FB-1001/500)
Puromycin (InvivoGen, catalog number: ant-pr-1) or equivalent
Reduced Serum media Opti-MEMTM (Thermo Fisher Scientific, GibcoTM catalog number: 31985047)
GeneJuice® Transfection Reagent (Merck Millipore, catalog number: 70967)
Phosphate-buffered saline (PBS) (Sigma, catalog number: D8537) or equivalent
Cell dissociation buffer Enzyme-free PBS-based (Thermo Fisher Scientific, GibcoTM catalog number: 13151014) to avoid surface cleavage of both hACE2 and TMPRSS2
Polybrene transfection reagent 10 mg/ml (Merk, catalog number: TR-1003-G)
Viability dye (Thermo Fisher Scientific, eBioscienceTM Fixable Viability Dye eFluorTM 780, catalog number: 65086514) or equivalent
Equipment
Class II biosafety cabinet (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraSafeTM 2030i)
Water bath (Avantor, VWB2 series, VWB2 2, catalog number: 462-0554) or equivalent
Incubator (Eppendorf, model: CellXpert® C170) or equivalent
Pipettes (Eppendorf Research® plus, P2, P20, P200, and P1000) or equivalent
Cell counter (Chemometec, Automated cell Analyzer NucleoCounter® NC-250TM) with Solution 18 AO.DAPI (Chemometec, catalog number: 910-3018) or equivalent
Plate centrifuge (Thermo Fisher Scientific, model: Heraeus Multifuge X3R) or equivalent
Flow cytometer equipped with a blue and red laser
This protocol utilized a BD LSRFortessaTM X-20 cell analyser but is not restricted to this equipment. In the absence of a flow cytometer, a fluorescent microscope fitted with a 488 nm excitation laser can be used.
Software
FlowJoTM Software, version 10.7.1 (BD Bioscience)
An alternative software for Flow Cytometry analysis can be used.
Procedure
Spike LV production by transient transfection
Day 1: Cell seeding
Seed 2.0 × 106 cells HEK293T/17 cells per 10 cm plate in 10 ml of culturing media supplemented with 2 mM L-glutamine and 10% FCS aiming for 80% confluence on day of transfection.
Day 3: Transfection
Prepare two sterile 1.5 ml microcentrifuge tubes labeled Tube 1 and Tube 2.
Add 5.56 µg of pCMVR8.74, 2.77 µg of pCDNA-SARS-CoV-2-S(D614G)ΔERR, and 4.17 µg of pCCL.SFFV.eGFP, for a total of 12.5 µg of DNA, to Tube 1 (equivalent to 2:1:1.5 plasmid ratios for scalable production).
Prepare the transfection mixture in Tube 2, as shown in Table 1.
Table 1. Transient transfection mixture.
Reagents μl/10 cm plate Gene Juice 30 OptiMEM® 470 Mix GeneJuice/OptiMEM® transfection mixture using a pipette and incubate for 5 min at room temperature.
Pipette 500 µl of transfection mixture into tube containing the plasmid constructs (Tube 1) and gently mix solution five times using the same pipette, avoiding the formation bubbles.
Incubate solution for 15 min at room temperature.
Pipette the DNA/GeneJuice/OptiMEM solution onto HEK293T/17 cells in a dropwise manner throughout the total surface of the plate.
Place plate in an incubator at 37 °C with 5% CO2 for 16-18 h.
Carefully remove the culture media from plate 16-18 h post-transfection.
Slowly add 7 ml of fresh culturing media supplemented with 2 mM L-glutamine and 10% FCS by pipetting media to one side of the plate to avoid detachment of cells.
Place plate back in the incubator at 37 °C with 5% CO2 for 16-18 h.
Harvest the viral supernatant containing the lentiviral vector pseudotyped with SARS-CoV-2-S(D614G)ΔERR into a 15 ml falcon previously chilled by either placing it in the fridge or on wet ice for 10 min.
Centrifuge at 400 × g for 5 min to remove cellular debris.
Clarify viral supernatant with 0.45 µm microfiltration to remove viral aggregates.
To avoid multiple freeze-thaw cycles, aliquot viral supernatants into 0.2-0.5 ml aliquots in sterile 1.5 ml microcentrifuge tubes with screw caps.
For later use, snap-freeze vials using dry ice and ethanol before storing them at -80 °C.
For immediate use, store vials on ice until needed.
Spike-LV titration by infectivity assay
Day 1:
Seeding permissive cells in a 24-well plate:
Pre-warm IMDM containing 10% FCS, 1% GlutaMAX, and 1 µg/ml Puromycin. This will be referred to as culturing media.
Harvest stable HEK293T/17-hACE2+TMPRSS2+ cells previously thawed and cultured for at least two passages (with the addition of 1 µg/ml Puromycin from the first passage after thawing) (Figure S1) as follows:
Remove and discard cellular supernatant.
Gently wash cells by pipetting 5 ml of PBS to remove any excess culturing media.
Add 3 ml of Cell dissociation buffer Enzyme-free PBS-based to the flask and incubate for 5 min.
Gently tap the sides of the flask to prompt cell detachment.
Add 7 ml of culturing media to flask, collect cells, and pipette into a new falcon.
Centrifuge at 400 × g for 5 min.
Discard supernatant and flick falcon with finger to gently dislodge cellular pellet.
Resuspend the cells by adding 5 ml of culturing media.
Determine cell density:
Passage cells by platting 1:6-1:8 of resuspended cells in a new T75 flask with 9 ml of culturing media, with a recommended minimum seeding of 3.3 × 104 cells/cm2.
Proceed to Step 1c for titration cell seeding.
Determine cell density using an Automated cell Analyzer NucleoCounter® NC-250TM.
Resuspend cells at 5.0 × 104cells/ml in 8 ml per viral vector supernatant to be quantified by titration.
Add 0.88 µl/ml of polybrene to resuspended cells; the final volume in each well will be 1.1 ml, leading to a final concentration of 8 µg/ml of polybrene.
Mix the resuspended cells and plate 1 ml/well in a total of seven wells (six wells for vector infection and one well for a no-vector control).
Add 1 ml of PBS to remaining unoccupied wells.
Preparing Spike LV dilutions for infection of permissive cells:
If using frozen Spike-LV, thaw in a water bath set at 37 °C and immediately use. If using fresh Spike-LV placed on ice, mix viral vectors by pipetting a few times, avoiding the formation of bubbles.
In a V-bottom 96-well plate, prepare the following 1:4 serial dilutions of Spike-LV in pre-chilled culture media for six points:
Pipette 200 µl of Spike-LV into one well in the V-bottom plate (e.g., well A1).
Pipette 150 µl of pre-chilled complete IMDM into five every other well in V-bottom plate (e.g., wells A3, A5, A7, A9, and A11).
Tip: Using every other well on the plate allows moving between 96-well plates and 24-well plates using a multichannel pipette (see Figure 1).
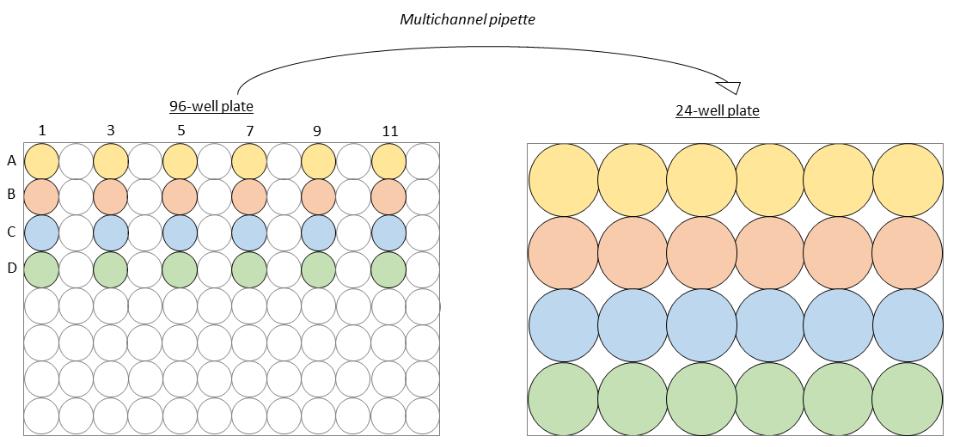
Figure 1. Transfer between 96-well and 24-well platesMove 50 µl from the first well (A1) containing Spike-LV into the subsequent well (A3).
Gently mix with a pipette ten times, being careful to avoid the formation of bubbles.
Move onto the third well (A5) and continue the serial dilution until reaching the 6th well (A11).
Using a multichannel with every other tip occupied, pipette 100 μl of vector dilutions to each well of HEK293T/17-hACE2+TMPRSS2+ cells already plated in a 24-well plate (Figure 1).
Centrifuge the plate at 1,000 × g for 20 min to enhance transduction by spinoculation.
Place plate back in the incubator at 37 °C with 5% CO2 for 72 h.
Discard culture media by flicking 24-well plate into a laboratory box that can be subsequently disinfected or a laboratory sink if the correct risk assessments are in place. Blot the plate on paper to remove excess liquid.
Add 200 μl/well of cell dissociation buffer and incubate for 10 min.
Gently tap the plate to detach cells.
Add 400 μl of PBS/well, resuspend cells by pipetting, and harvest 200 μl into a new V-bottom 96-well plate.
Tip: Using every other well on the plate allows moving between 96-well plates and 24-well plates using a multichannel pipette (Figure 1).
Centrifuge the plate at 400 × g for 5 min.
Discard supernatant by quickly flicking plate in an appropriate manner (see step 1) and blot on paper to remove excess liquid.
Add 250 μl PBS/well and centrifuge plate at 400 × g for 5 min.
In the meantime, prepare a master mix for Fixable Viability Dye eFluorTM 780 (0.1 µl + 99.9 µl PBS/well).
Discard supernatant by quickly flicking plate in an appropriate manner (see step 1) and blot on paper to remove excess liquid.
Resuspend cells in 100 µl /well staining master mix.
Incubate at room temperature for 30 min in the dark.
Add 150 μl PBS/well and centrifuge plate at 400 × g for 5 min.
Discard PBS, blot plate, and resuspend cells in 50 μl of PBS using P200 tips.
Acquire plate on flow cytometer with the following bandpass filters:
RL2 780/60 for Fixable Viability Dye eFluorTM 780.
BL1 530/60 for fluorescent marker of transduction, eGFP.
Note: If the user is unfamiliar with flow cytometric analysis of cells, it is advisable that a trained flow cytometry operator assists from this step onward.
Data analysis
FlowJo analysis
The gating strategy to identify the percentage of eGFP-positive population is depicted in Figure 2A, starting with the live cell gate, followed by the forward versus side scatter gate, and the singlets identification. Singlets are then plotted as histograms to identify the percentage of eGFP-positive cells (Figure 2B).
Determine the percent of transduced cells in each well
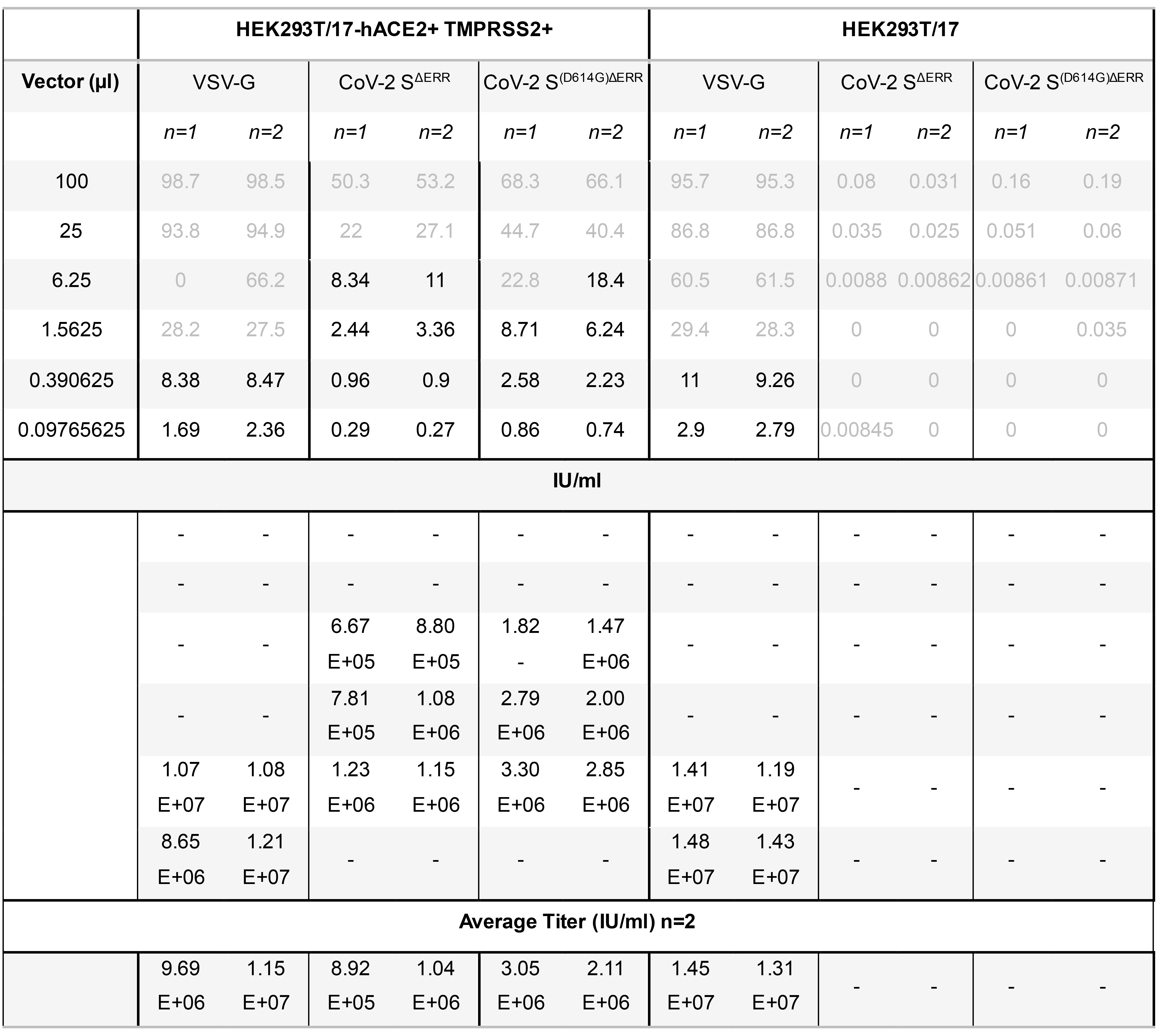
When calculating titers, only transduction efficiencies of less than 20% of transduced cells should be considered. This method assumes one integration per cell. Using >20% risks counting cells with multiple integration sites which leads to an underestimation of the titer. Transduction efficiencies between 0.5% and 20% were used to compare vector production between a control LV incorporating a Vesicular Stomatitis virus glycoprotein (VSV-G) and LV pseudotyped with CoV-2 SΔERR, with or without the prevalent D614G mutation. The D614G mutation was found to increase the titer by approximately 60% (Figure 2C). Titration calculations are presented in Table 1 as an example.
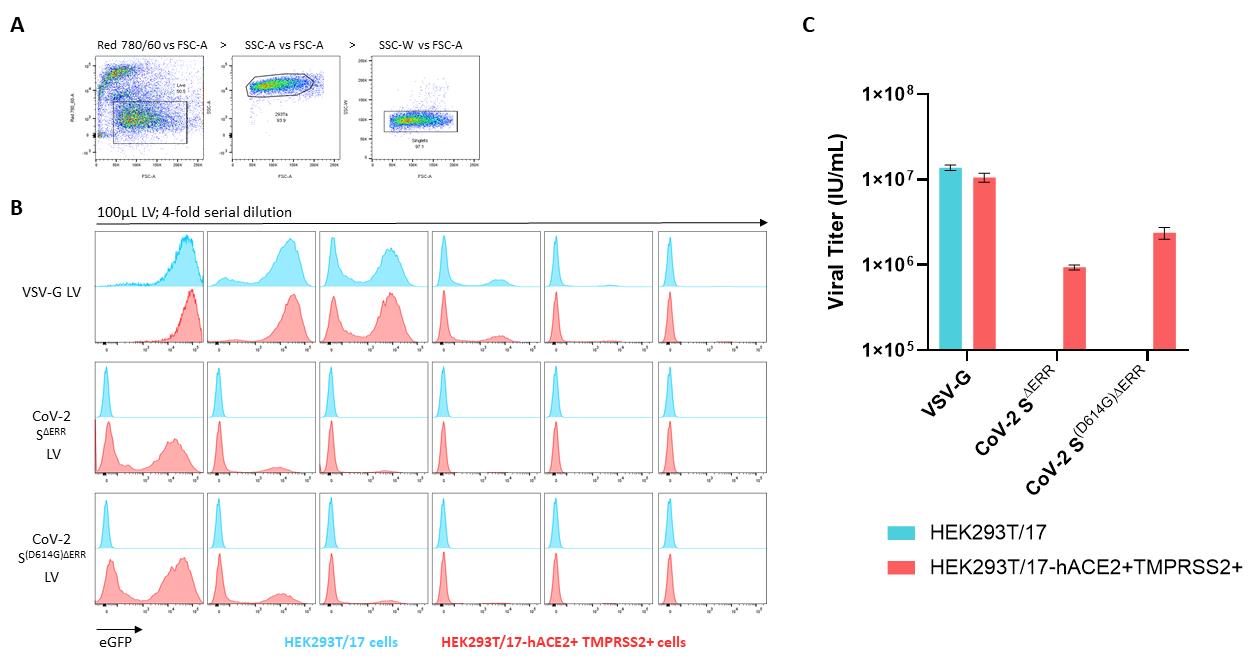
Figure 2. Flow cytometric analysis of pseudotyped lentiviral vectors on stable HEK293T/17-hACE2+TMPRSS2+. (A) Gating strategy starting with live (eF780) vs. FSC-A to identify live cells, followed by SSC-A vs. FSC-A to identify HEK293T/17 cells, and by SSC-W vs. FSC-A for singlets identification. (B) Transiently produced VSV-G LV, SΔERR CoV-2 LV, and S(D614G)ΔERR CoV-2 LV were serially diluted from 100 µl in a 4-fold dilution for six points and added on control HEKT293T/17 cells (blue population) and CoV-2 S permissive HEK293T/17-hACE2+TMPRSS2+ cells (red population) in the presence of 8 µg/ml of polybrene. Staggered histograms of transduced populations are presented. (C) Graph representing viral titers of the three pseudotyping glycoproteins quantified by an infectivity assay on HEK293T/17 and HEK293T/17-hACE2+TMPRSS2 cells. Data presented are ±SD of duplicate determinations.Calculate the functional titer as infectious units per ml (IU/ml) using the following equation:
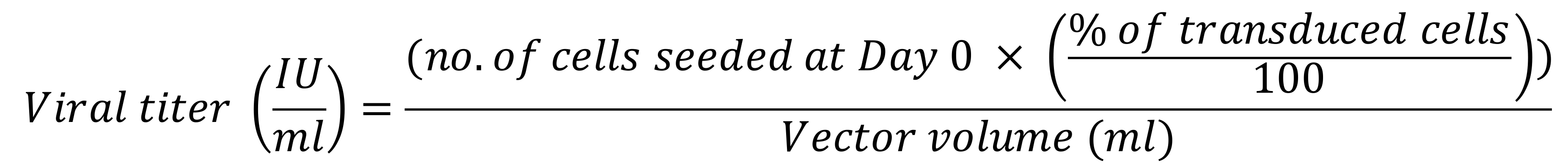
Note: For a more accurate titer, take the average of multiple dilutions.
Table 2. Calculation of lentiviral vector titers. Viral titer calculation of lentiviral vector pseudotyped with VSV-G, CoV-2 SΔERR LV, and CoV-2 S(D614G)ΔERR infected on both HEK293T/17 cells and HEK293T/17-hACE2+TMPRSS2+ cells. Transduction efficiencies higher than 20% and lower than 0.5% are highlighted in grey font and omitted from titer calculation. Average titers are presented from duplicate determinations.
Acknowledgments
The authors acknowledge Dr Nigel Temperton (Medway School of Pharmacy, The University of Kent) for the pCDNA-SARS-CoV-2-Spike plasmid expressing the full-length codon-optimised Spike glycoprotein. E. M. B was funded by the Coalition for Epidemic Preparedness Innovations (CEPI).
Competing interests
The authors declare no competing interests.
References
- Carnell, G., Grehan, K., Ferrara, F., Molesti, E. and Temperton, N. (2017). An Optimized Method for the Production Using PEI, Titration and Neutralization of SARS-CoV Spike Luciferase Pseudotypes. Bio-protocol 7(16): e2514.
- Chen, W. H., Strych, U., Hotez, P. J. and Bottazzi, M. E. (2020). The SARS-CoV-2 Vaccine Pipeline: an Overview. Curr Trop Med Rep: 1-4.
- Ferrari, M., Mekkaoui, L., Ilca, F.T., Akbar, Z., Bughda, R., Lamb, K., Ward, K., Parekh, F., Karattil, R., Allen, C., Wu, P., Baldan, V., Mattiuzzo, G., Bentley, E.M., Takeuchi, Y., Sillibourne, J., Datta, P., Kinna, A., Pule, M., Onuoha, S.C. (2021). Characterisation of a novel ACE2-based therapeutic with enhanced rather than reduced activity against SARS-CoV-2 variants. J Virol: JVI0068521.
- Fukushi, S., Mizutani, T., Saijo, M., Matsuyama, S., Miyajima, N., Taguchi, F., Itamura, S., Kurane, I. and Morikawa, S. (2005). Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J Gen Virol 86(Pt 8): 2269-2274.
- Grehan, K., Ferrara, F. and Temperton, N. (2015). An optimised method for the production of MERS-CoV spike expressing viral pseudotypes. MethodsX 2: 379-384.
- Hoffmann, M., Kleine-Weber, H. and Pohlmann, S. (2020a). A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol Cell 78(4): 779-784 e775.
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Kruger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N. H., Nitsche, A., Muller, M. A., Drosten, C. and Pohlmann, S. (2020b). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181(2): 271-280 e278.
- Ju, B., Zhang, Q., Ge, J., Wang, R., Sun, J., Ge, X., Yu, J., Shan, S., Zhou, B., Song, S., Tang, X., Yu, J., Lan, J., Yuan, J., Wang, H., Zhao, J., Zhang, S., Wang, Y., Shi, X., Liu, L., Zhao, J., Wang, X., Zhang, Z. and Zhang, L. (2020). Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584(7819): 115-119.
- Korber, B., Fischer, W. M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., Hengartner, N., Giorgi, E. E., Bhattacharya, T., Foley, B., Hastie, K. M., Parker, M. D., Partridge, D. G., Evans, C. M., Freeman, T. M., de Silva, T. I., Sheffield, C.-G. G., McDanal, C., Perez, L. G., Tang, H., Moon-Walker, A., Whelan, S. P., LaBranche, C. C., Saphire, E. O. and Montefiori, D. C. (2020). Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182(4): 812-827 e819.
- Letko, M., Marzi, A. and Munster, V. (2020). Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5(4): 562-569.
- Nie, J., Li, Q., Wu, J., Zhao, C., Hao, H., Liu, H., Zhang, L., Nie, L., Qin, H., Wang, M., Lu, Q., Li, X., Sun, Q., Liu, J., Fan, C., Huang, W., Xu, M. and Wang, Y. (2020). Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect 9(1): 680-686.
- Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., Guo, L., Guo, R., Chen, T., Hu, J., Xiang, Z., Mu, Z., Chen, X., Chen, J., Hu, K., Jin, Q., Wang, J. and Qian, Z. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11(1): 1620.
- Peacock, T. P., Goldhill, D. H., Zhou, J., Baillon, L., Frise, R., Swann, O. C., Kugathasan, R., Penn, R., Brown, J. C., Sanchez-David, R. Y., Braga, L., Williamson, M. K., Hassard, J. A., Staller, E., Hanley, B., Osborn, M., Giacca, M., Davidson, A. D., Matthews, D. A. and Barclay, W. S. (2021). The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. doi: 10.1038/s41564-021-00908-w.
- Pinto, D., Park, Y. J., Beltramello, M., Walls, A. C., Tortorici, M. A., Bianchi, S., Jaconi, S., Culap, K., Zatta, F., De Marco, A., Peter, A., Guarino, B., Spreafico, R., Cameroni, E., Case, J. B., Chen, R. E., Havenar-Daughton, C., Snell, G., Telenti, A., Virgin, H. W., Lanzavecchia, A., Diamond, M. S., Fink, K., Veesler, D. and Corti, D. (2020). Structural and functional analysis of a potent sarbecovirus neutralizing antibody. bioRxiv 2020.04.07.023903.
- Quinlan, B.D., Mou, H., Zhang, L., Guo, Y., He, W., Ojha, A., Parcells, M.S., Luo, G., Li, W., Zhong, G., Choe, H. and Farzan, M. (2020). The SARS-CoV-2 Receptor-Binding Domain Elicits a Potent Neutralizing Response Without Antibody-Dependent Enhancement. (SSRN Scholarly Paper No. ID 3575134). Social Science Research Network, Rochester, NY.
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T. and Veesler, D. (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181(2): 281-292 e286.
- Wu, Y. (2020). Strong evolutionary convergence of receptor-binding protein spike between COVID-19 and SARS-related coronaviruses. bioRxiv 2020.03.04.975995.
- Yan, K. X., Tan, W. J., Zhang, X. M., Wang, H. J., Li, Y. and Ruan, L. (2007). Development and application of a safe SARS-CoV neutralization assay based on lentiviral vectors pseudotyped with SARS-CoV spike protein. Bing Du Xue Bao 23(6): 440-446.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mekkaoui, L., Bentley, E., Ferrari, M., Lamb, K., Ward, K., Karattil, R., Akbar, Z., Bughda, R., Sillibourne, J., Onuoha, S., Mattiuzzo, G., Takeuchi, Y. and Pule, M. (2021). Optimised Method for the Production and Titration of Lentiviral Vectors Pseudotyped with the SARS-CoV-2 Spike. Bio-protocol 11(16): e4194. DOI: 10.21769/BioProtoc.4194.
Category
Molecular Biology > Protein > Detection
Microbiology > Microbe-host interactions > Virus
Biochemistry > Protein > Synthesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








