- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Enrichment of Vascular Fragments from Mouse Embryonic Brains for Endothelial Cell Analysis
Published: Vol 11, Iss 12, Jun 20, 2021 DOI: 10.21769/BioProtoc.4058 Views: 3419
Reviewed by: Pilar Villacampa AlcubierreAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2243 Views

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1343 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1475 Views
Abstract
Endothelial cells in the brain interact with other cell types, forming the blood-brain barrier. This barrier controls the movement of solutes into and out of the brain, regulating pathophysiological processes and drug delivery to the brain. Common isolation methods used to study these cells during embryonic development involve enzymatic treatment and cell sorting using specific markers. This process modifies the cell state and produces minute amounts of sample. Here, we describe a protocol for the enrichment of vascular cells from embryonic brains based on dextran separation. In this method, the brain is lightly disrupted with a pestle and then resuspended in a dextran solution. Low-speed centrifugation permits the separation of the parenchymal and vascular fractions. Further centrifugation steps improve fractionation. This method is simple and fast and produces enough sample for biochemical assays.
Graphic abstract:
Purification of vascular fragments from an embryonic brain
Background
Endothelial cells in the brain form a specialized barrier that limits and controls the passage of large and small molecules from the bloodstream into the brain (Zlokovic, 2008). In their natural niche, brain endothelial cells interact closely with several cell types, including pericytes, glia, and immune cells, which are integral to brain homeostasis (Armulik et al., 2005; Gavins et al., 2007). The study of brain endothelial cell biology has benefited greatly from the use of procedures to isolate either endothelium or vascular complexes, which include endothelial cells, pericytes, and glial contacts (radial glia, oligodendrocytes, and astrocyte endfeet). The former is normally achieved by fluorescence-activated cell sorting (FACS) isolation (Daneman et al., 2010), whereas the latter is done by centrifugation in a dextran solution (Yousif et al., 2007). FACS-based isolation can alter the properties of endothelial cells and is expensive and inefficient, leading to the isolation of very few cells that may not reflect their native state. These issues are more evident when obtaining samples from embryonic brains, where the sample is extremely limited, and fewer than 5000 endothelial cells are typically obtained from one brain. This number is adequate for transcriptional analyses using specialized kits (Santander et al., 2020) but is insufficient for biochemical assays. Conversely, dextran-based isolation of vascular complexes has not been applied to embryonic brains because the standard protocol requires a much bigger sample obtained from single mouse embryos. The ability to obtain enough sample for biochemical assays from single embryonic brains is necessary for the study of developmental vascular diseases, such as the Fowler syndrome (Santander et al., 2020). Here, to overcome these sample size issues, we describe the adaptation of the paradigm of dextran-based isolation of vascular fragments to individual mouse embryo brains. We demonstrate the utility of vascular fragment isolation to measure cholesterol content in embryonic brain blood vessels, an assay not feasible using FACS-isolated endothelial cells.
Materials and Reagents
15 ml conic bottom tube
DuraSeal laboratory film (Sigma, catalog number: D3172)
Polypropylene 1.5 ml tubes
Assay plate, 96-well, black
Assay plate, 96-well, clear
PVDF membrane (Thermo, catalog number: 88520)
Culture plate
Female and male mice
PBS 10× (Apex, catalog number: 20-134)
Dextran (70 kDa Highly hygroscopic, should be maintained in an inert atmosphere) (TCI, catalog number: D1449)
IGEPAL (Sigma, catalog number: I8896)
Sodium dodecyl sulfate (Sigma, catalog number: L3771)
Protease inhibitor cocktail (Thermo, catalog number: 78429)
MilliQ water
BCA Protein Assay kit (Thermo, catalog number: 23225)
Methanol (Merck, catalog number: 1060091000)
Chloroform (Merck, catalog number: 1070242500)
Amplex Red Cholesterol Assay Kit (Thermo, catalog number: A12216)
Protein loading buffer 4× (Bio-Rad, catalog number: 1610747)
2-mercaptoethanol (Sigma, catalog number: M6250)
Buffer Tris-Glycine 10× (Apex, catalog number: 18-238B)
Pre-cast 10% polyacrylamide gel (Bio-Rad, catalog number: 4561033)
Powdered skim milk
TBST 10× (Apex, catalog number: 18-235B)
Rabbit anti-ERG (Abcam, catalog number: ab92513)
Rabbit anti-PECAM1 (Santa Cruz, catalog number: sc-1506)
Rabbit anti-PAX6 (Millipore, catalog number: AB2237)
Rabbit anti-TBR1 (Abcam, catalog number: ab31940)
Mouse anti-ACTINB (R&D Systems, catalog number: MAB8929)
Donkey anti-rabbit IGG (Thermo, catalog number: A16029)
Donkey anti-mouse IGG (Thermo, catalog number: A16011)
Chemiluminescence kit (Bio-Rad, catalog number: 1705061)
PBS 1× (see Recipes)
18% Dextran (see Recipes)
Lysis buffer (see Recipes)
Running buffer (see Recipes)
Transfer buffer (see Recipes)
TBST 1× (see Recipes)
Equipment
Dumont #5 fine forceps (Fine Science Tools, catalog number: 11251-30)
Tube pestle (Sigma, catalog number: Z359947)
CO2 euthanasia chamber
Dissection microscope
Refrigerated microcentrifuge
Heating block
Fume hood
Gel documentation system
Software
LibreOffice Calc
R
Procedure
Embryo brain dissection
Breed female and male mice in a 2:1 or 1:1 ratio.
Check females for the presence of a vaginal plug every day within the first hour of the light cycle.
Separate females with a plug to avoid re-plugging and register that morning as embryonic day (E) 0.5.
On day E15.5, euthanize females following institutional guidelines. We use a combination of CO2 asphyxiation and cervical dislocation. See Note 1.
Remove the uteri, place them in PBS 1×, and separate each implantation site. Care should be taken to maintain the yolk sac intact to minimize tissue damage.
Under a dissection microscope, dissect the embryo free of extraembryonic tissues. If genotyping is required, cut the tip of the tail or a paw and place it in a tube. See Note 2.
Cut the embryo head and peel the skin and meningeal membranes covering the brain using Dumont #5 fine forceps. See Note 3 and Figure 1.
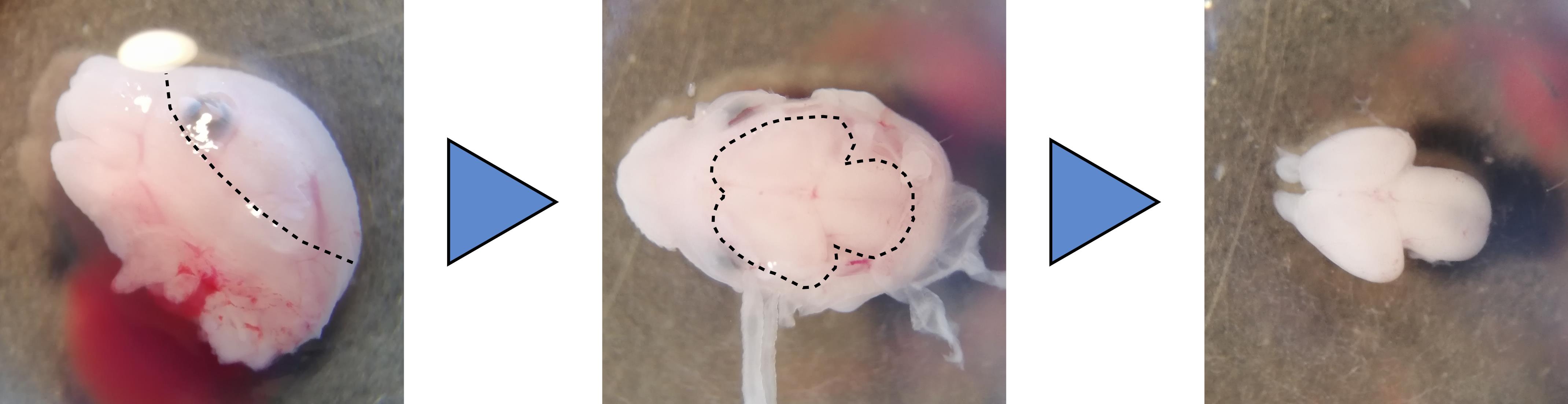
Figure 1. Dissection of a mouse embryo brain. The head skin is peeled away along the dotted line using Dumont #5 forceps (Left). The meningeal layers covering the brain are removed carefully to expose the brain (Center, dotted line). Pushing the brain gently from the side releases the whole organ from the head (Right).Remove the brain from the head by cutting the rhombencephalon at the base of the skull and gently pushing the forebrain from the side.
Place the brain in 1 ml of ice-cold PBS in a 1.5 ml polypropylene tube.
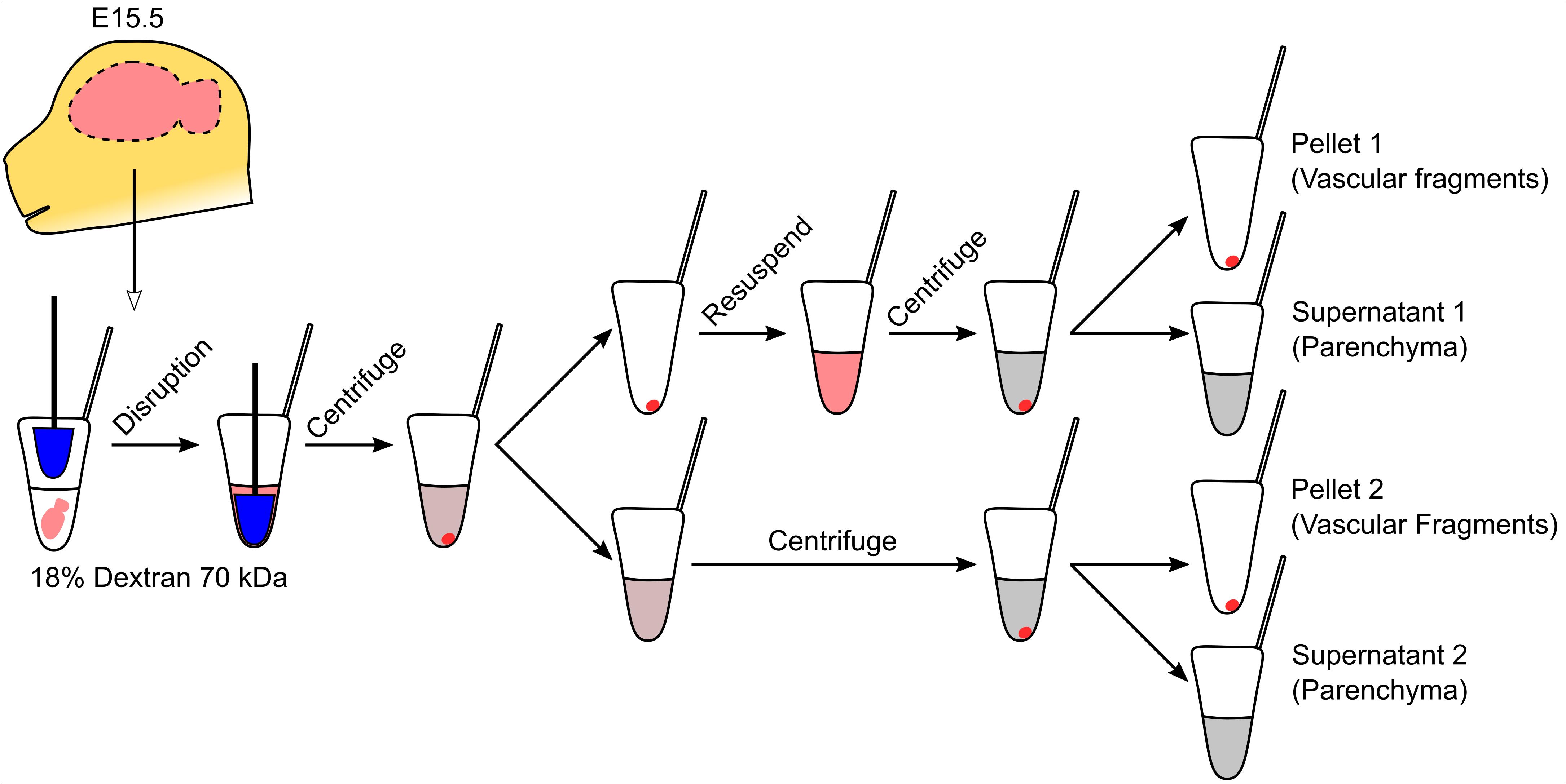
Vascular fragment isolation (all steps should be performed on ice)
Disrupt brains with five strokes using a sterile 1.5 ml tube pestle.
Centrifuge at 1,500 × g for 20 min at 4°C.
During centrifugation, prepare enough 18% dextran solution in PBS to individually process each of the brains collected.
Discard the supernatant and resuspend the pellet in 500 μl of 18% dextran. See Note 4.
Centrifuge at 1,500 × g for 20 min at 4°C.
Transfer the supernatant to another tube. See Note 5.
Resuspend the pellet in 500 μl of 18% dextran.
Centrifuge both fractions at 1,500 × g for 20 min at 4°C. Pellets are the vascular fragments.
Transfer the supernatant to a different tube and add 1 ml of PBS.
Centrifuge the diluted supernatants at 10,000 × g for 10 min at 4°C and discard the supernatant. The pellets are the parenchymal fractions.
Store vascular and parenchymal fractions at < -70°C until use.
Western blotting for the evaluation of enrichment of fractions
Add 100 μl of lysis buffer directly to the pellet on ice. Care should be taken to maintain the pellet frozen until the lysis buffer is added by keeping the tubes on dry ice or liquid nitrogen.
Lyse the sample by pipetting up and down until the lysate looks homogeneous. See Note 6.
Pool pellets corresponding to the same fractions of the same sample by transferring all the lysate from the first tube to the second. Lyse the second pellet by pipetting. See Note 7.
Transfer 10 μl of lysate to a different 1.5 ml tube for protein quantification. Keep at < -70°C.
Repeat Steps C1-C4 for each sample to be analyzed in parallel. Always process controls alongside experimental samples.
Add 2-mercaptoethanol to loading buffer to a concentration of 5%.
Mix an aliquot of 15 μg of protein with 1/3 volume of loading buffer with 2-mercaptoethanol and heat at 98°C for 5 min.
Load into a 10% polyacrylamide gel and run for the appropriate time to resolve proteins in the range of 30 kDa and 150 kDa in Running buffer.
Incubate a PVDF membrane in methanol for 5 min for activation.
Transfer proteins into the activated PVDF membrane at 300 mA for 1 h in Transfer buffer.
Block the membrane by incubating 2 h in 5% skim milk, 0.01% Tween 20 in TBS 1×.
Incubate the membrane with the primary antibody diluted 1:2,000 in blocking buffer.
Wash the membrane three times for 10 min in 0.01% Tween 20 in TBS 1×.
Incubate with the secondary antibody diluted 1:10,000 in blocking buffer.
Wash the membrane three times for 10 min in 0.01% Tween 20 in TBS 1×.
Mix the solutions from the chemiluminescence kit and cover the membrane for 1 min.
Blot excess solution and image in a gel documentation system.
A representative blot demonstrating the enrichment of endothelial markers in the vascular fraction and of a neuronal progenitor marker in the parenchymal fraction is shown in Figure 2.
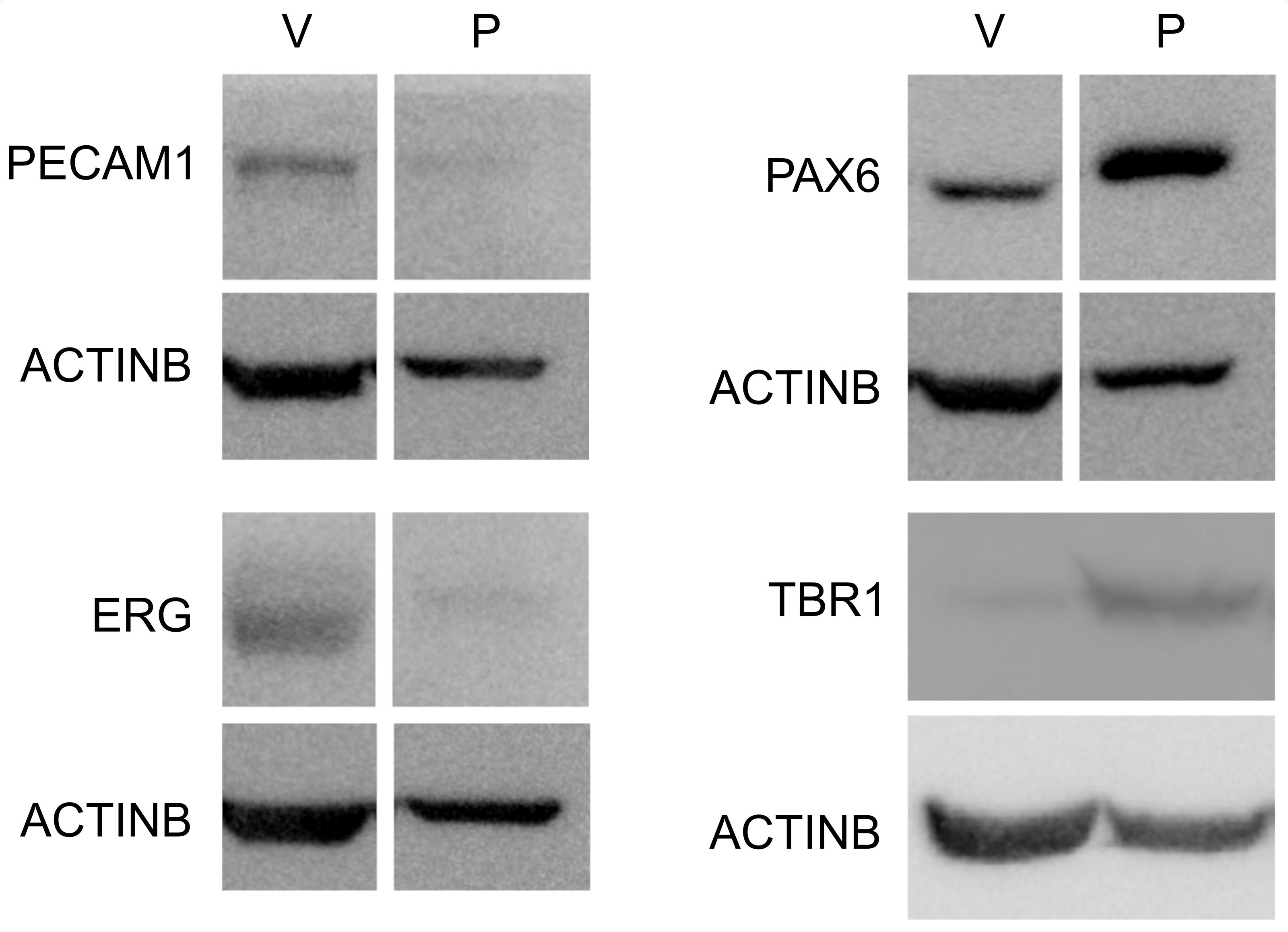
Figure 2. Enrichment of vascular and parenchymal fractions. Western blot analysis of endothelial (PECAM1, ERG) and neuronal progenitor (PAX6, TBR1) markers in the vascular (V) and parenchymal (P) fractions. Note that the PAX6 signal is detected in vascular fragments, representing PAX6 expression in brain endothelial cells (Vasudevan et al., 2008) and/or vascular adherent neuro/glial progenitors (Tan et al., 2016).
Lipid extraction
Add 100 μl of lysis buffer directly to the pellet on ice. Care should be taken to maintain the pellet frozen until lysis buffer is added by keeping the tubes on dry ice or liquid nitrogen.
Lyse the sample by pipetting up and down until the lysate looks homogeneous. See Note 6.
Pool pellets corresponding to the same fractions of the same sample by transferring all the lysate from the first tube to the second. Lyse the second pellet by pipetting. See Note 7.
Transfer 10 μl of lysate to a different 1.5 ml tube for protein quantification. Keep at < -70°C.
Repeat Steps D1-D4 for each sample to be analyzed in parallel. Always process controls alongside experimental samples.
In a fume hood, add 400 μl of methanol to the 90 μl of lysate, followed by 800 μl of chloroform. See Note 8.
Seal tubes with DuraSeal laboratory film. See Note 9.
Vortex each tube for 1 min to mix both phases well.
Heat at 60°C for 30 min using a heating block or bath.
Vortex for 1 min.
Incubate overnight at 4°C to allow phases to separate.
Centrifuge at 1,500 × g for 20 min at 4°C.
Transfer 400 μl of the hydrophobic (lower) phase into a new tube. To avoid contaminating the organic phase with the aqueous one, push some air into the pipette tip before introducing it into the solution, and release it in the upper part of the lower phase.
Transfer 50 μl of organic phase into a new tube and evaporate under an inert atmosphere. After evaporation, no crystallization should be visible. Two tubes should be prepared in this manner for each sample to measure cholesterol in duplicate.
Cholesterol measurement
Prepare all reagents in the Amplex Red Cholesterol Assay Kit, following the manufacturer’s instructions.
Add 50 μl of Reaction buffer 1× to the tubes containing the evaporated organic phase.
Vortex for 1 min. See Note 10.
Transfer to a black 96-well plate.
Construct a cholesterol standard curve following the manufacturer’s instructions. Transfer 50 μl to the black 96-well plate.
Set up the reaction mixture as described by the manufacturer and add 50 μl to each well containing the samples and the curve.
Incubate at 37°C for 30 min.
Measure the fluorescence with excitation at 560 nm and emission at 590 nm.
Protein measurement
Thaw sample aliquots on ice and prepare 100 μl of a 10-fold dilution by adding 90 μl of MilliQ water to each tube.
Construct a BSA curve by diluting the BSA standard as instructed by the manufacturer.
Transfer 25 μl of each sample and curve point to a clear 96-well plate.
Prepare enough reaction mixture for all samples and the curve by mixing Buffer A and Buffer B in a 50:1 ratio.
Add 200 μl of reaction mixture to each well.
Incubate at 37°C for 30 min.
Measure the absorbance at 562 nm.
Data analysis
Use the LibreOffice Calc software to estimate cholesterol and protein concentration in each sample by interpolating into the appropriate standard curve. These values correspond to cholesterol content in 50 μl of extract and proteins in 1 μl of lysate (considering the 10-fold dilution).
Multiply cholesterol levels by 16 to obtain levels in 800 μl of chloroform and protein levels by 50 to obtain the amount in the entire lysate.
Express cholesterol levels as μg of cholesterol per μg of protein to compare different samples.
Compare groups in the R statistical environment. When two groups are compared, a t-test is performed; if more than two groups are being analyzed, use ANOVA with a Tukey’s post-test. In Figure 3, we compare cholesterol levels in vascular and parenchymal fractions. Since both fractions come from the same sample, a paired t-test was performed.
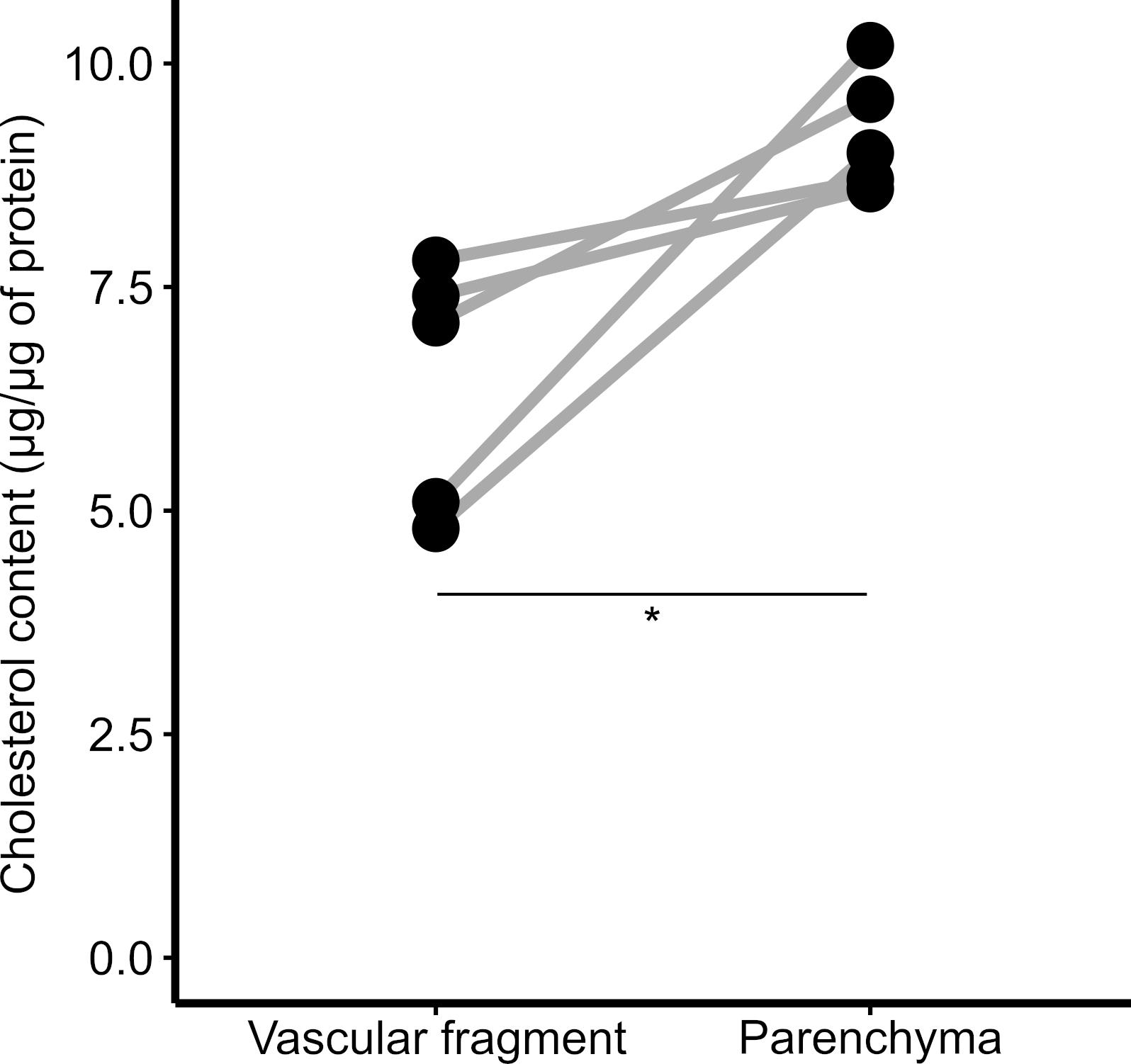
Figure 3. Cholesterol content in brain fractions. Vascular and parenchymal fractions were isolated using dextran-based centrifugation, and the cholesterol content in each fraction was determined. *P < 0.05; paired t-test.
Notes
E15.5 is the earliest time point at which this protocol has worked in our hands. It is likely due to the fact that the procedure relies on the high density of the endothelium-pericyte-astrocyte endfeet complex, which begins to mature at around E15.5.
Since genotyping cannot be done in a short time frame, all embryos need to be processed for vascular fragment isolation, even if only some of the embryos are expected to carry useful genotypes. After genotyping, we use the samples from embryos with required genotypes without excluding any sample.
The easiest way is to put the head sideways is to grab the skin just above the presumptive ear with one forceps and begin peeling the rest of the skin with the other forceps. Just below the skin and covering the brain are the translucent meningeal membranes. To remove them, pinch carefully between forebrain hemispheres with one forceps and peel away the membranes with the other (Figure 1).
The tissue will look gelatinous but evenly distributed in the dextran solution. In our hands, it looks more like a homogenate than a lightly disrupted tissue.
The parenchymal fraction will remain in the supernatant, whereas the vascular fragments will be pelleted. Care should be taken to recover as much parenchyma as possible.
Lysis occurs rapidly after thawing the pellet in lysis buffer, but fibrous material may be present after lysis of the vascular fraction. We include this in the lipid extraction but remove it by centrifugation for other applications, such as Western blotting.
This strategy for pooling pellets can be used to mix several different samples without increasing the volume; this is useful if detection methods require more material than what is possible to obtain from one embryonic brain. When collecting samples for new applications, we always test the new assay using pools of 1-3 samples to determine the detection limit. In our hands, a sample from 1 embryonic brain is sufficient for cholesterol determination and Western blotting.
Polypropylene tubes may react with chloroform, especially when heated. If tubes change form, small glass tubes should be used instead. In our hands, polypropylene tubes normally withstand the conditions of this procedure, but glass tubes may be preferable.
DuraSeal is a solvent-resistant parafilm replacement that will prevent solvent evaporation during heating. Three layers should be applied to tightly seal the tube by covering the tube cap with a film square and twisting and recovering twice. When done correctly, no chloroform should leak when inverting the tube.
It is important to resuspend precipitated lipids quantitatively. We have used vortexing, but it is common to use a bath sonicator for 15 min. Both time and type of resuspension should be tested when setting up this procedure.
Recipes
PBS 1×
Dilute 100 ml of PBS 10× to 1 L with MilliQ water
18% Dextran
To prepare enough to process one brain, dissolve 9 mg of dextran 70 kDa in 500 μl of PBS 1×
Prepare enough volume to process all brains, considering an excess volume of 1 ml
Lysis buffer
50 mM Tris
150 mM NaCl
1% IGEPA
0.1% Sodium Dodecyl Sulfate
Protease inhibitors 1×
Running buffer
Mix 100 ml of Tris-Glycine 10× and 10 ml of SDS 10% and bring to 1 L with MilliQ water
Transfer buffer
Mix 100 ml of Tris-Glycine 10× and 200 ml of methanol and bring to 1 L with MilliQ water
TBST 1×
Dilute 100 ml of TBST 10× to 1 L with MilliQ water
Acknowledgments
N.S. is supported by AHA Postdoctoral fellowship 20POST35120371. This protocol is based on Boulay et al. (2015).
Competing interests
The authors declare that no competing interests exist.
Ethics
Animal procedures described here follow AVMA guidelines and have been approved by the Institutional Review Board of the University of California, San Francisco (AN177934 2019-2022).
References
- Armulik, A., Abramsson, A. and Betsholtz, C. (2005). Endothelial/pericyte interactions. Circ Res 97(6): 512-523.
- Boulay, A. C., Saubamea, B., Decleves, X. and Cohen-Salmon, M. (2015). Purification of Mouse Brain Vessels. J Vis Exp(105): e53208.
- Daneman, R., Zhou, L., Agalliu, D., Cahoy, J. D., Kaushal, A. and Barres, B. A. (2010). The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS One 5(10): e13741.
- Gavins, F., Yilmaz, G. and Granger, D. N. (2007). The evolving paradigm for blood cell-endothelial cell interactions in the cerebral microcirculation. Microcirculation 14(7): 667-681.
- Santander, N., Lizama, C. O., Meky, E., McKinsey, G. L., Jung, B., Sheppard, D., Betsholtz, C. and Arnold, T. D. (2020). Lack of Flvcr2 impairs brain angiogenesis without affecting the blood-brain barrier. J Clin Invest 130(8): 4055-4068.
- Tan, X., Liu, W. A., Zhang, X. J., Shi, W., Ren, S. Q., Li, Z., Brown, K. N. and Shi, S. H. (2016). Vascular Influence on Ventral Telencephalic Progenitors and Neocortical Interneuron Production. Dev Cell 36(6): 624-638.
- Vasudevan, A., Long, J. E., Crandall, J. E., Rubenstein, J. L. and Bhide, P. G. (2008). Compartment-specific transcription factors orchestrate angiogenesis gradients in the embryonic brain. Nat Neurosci 11(4): 429-439.
- Yousif, S., Marie-Claire, C., Roux, F., Scherrmann, J. M. and Decleves, X. (2007). Expression of drug transporters at the blood-brain barrier using an optimized isolated rat brain microvessel strategy. Brain Res 1134(1): 1-11.
- Zlokovic, B. V. (2008). The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron 57(2): 178-201.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Santander, N. and Arnold, T. D. (2021). Enrichment of Vascular Fragments from Mouse Embryonic Brains for Endothelial Cell Analysis. Bio-protocol 11(12): e4058. DOI: 10.21769/BioProtoc.4058.
Category
Biochemistry > Other compound
Neuroscience > Development
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








