- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Detailed Protocol for Preparing Millimeter-sized Supergiant Liposomes that Permit Efficient Eukaryotic Cell-free Translation in the Interior
Published: Vol 11, Iss 12, Jun 20, 2021 DOI: 10.21769/BioProtoc.4054 Views: 3078
Reviewed by: Ali Asghar KermaniKumiko OkazakiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Translating Ribosome Affinity Purification (TRAP) of Cell Type-specific mRNA from Mouse Brain Lysates
Catherine L. Salussolia [...] Mustafa Sahin
May 5, 2022 5423 Views

Human-rabbit Hybrid Translation System to Explore the Function of Modified Ribosomes
Eriko Matsuura-Suzuki [...] Shintaro Iwasaki
Jul 5, 2023 2315 Views
Abstract
Liposomes have been used as a pseudo cell membrane for encapsulating biomolecules and creating an artificial cell in the interior where biochemical reactions can occur. Among the several methods used to prepare biomolecule-encapsulating liposomes, the spontaneous emulsion transfer method is superior to others in that it allows us to readily prepare relatively large liposomes whose sizes are controlled (from micrometer- to millimeter-sized liposomes) without special equipment. However, conventional protocols for this method require liposomes to contain a considerably high concentration of sucrose (high-density solute), which severely inhibits gene expression, one of the most important biochemical reactions. Thus, we optimized the preparation conditions to develop a wheat germ extract (WGE)-based protocol that requires a much lower concentration of sucrose and has almost no effect on eukaryotic cell-free translation. Our protocol allows us to successfully prepare millimeter-sized, moderately stable, WGE-encapsulating liposomes in which WGE translation takes place efficiently. Since a broad range of genes derived from various types of organisms can be efficiently translated in a WGE-based translation system, liposomes prepared using our protocol would be useful as a versatile research tool for artificial cells.
Keywords: Artificial cellBackground
Liposomes are spherical vesicles composed mainly of double-layered phospholipids, similar to a living cell membrane; thus, they have been used as containers for biomolecules to construct artificial cells that allow for biochemical reactions (e.g., gene expression) in the interior separately from and/or in communication with the exterior (Walde et al., 2010).
Several methods are available for preparing liposomes that encapsulate biomolecules. In particular, a manual emulsion transfer method is useful for readily preparing relatively large liposomes with a diameter of more than 1 μm (called giant unilamellar vesicles, GUVs) without the need for special equipment such as an evaporator or a microfluidic device (Pautot et al., 2003; Noireaux et al., 2004). In this method, a water-in-oil emulsion that is covered with a phospholipid layer in an upper oil phase (containing phospholipids) is transferred into a lower water phase to be further covered with another phospholipid layer in the phase interface, thus forming a liposome.
Although a centrifuge is generally used for the phase transfer, an emulsion droplet can be transferred spontaneously just by adding a high-density solute (e.g., sucrose) without centrifugation, which often forces the formed liposome to burst (Hamada et al., 2008). Needless to say, the difference in density between the droplet and the water phase is key to the successful spontaneous phase transfer. This spontaneous method enables us to control the size of a liposome by adjusting the droplet volume, and surprisingly, to prepare a supergiant liposome with a diameter of more than 1 mm (called a supergiant unilamellar vesicle, SGUV), which can be observed by the naked eye without a microscope using a microliter-sized droplet (Kubatta et al., 2009).
However, conventional protocols for spontaneous emulsion transfer have the drawback that they require the inclusion of a considerably high concentration of sucrose (10-30% w/v) in the droplet that is to be spontaneously transferred. Such high amounts of sucrose severely inhibit gene expression, one of the most important biochemical reactions (Elani et al., 2015). For example, eukaryotic cell-free translation based on wheat germ extract (WGE) is suppressed by more than 80% in the presence of 10% w/v sucrose (Takahashi and Ogawa, 2020). Thus, we optimized the preparation conditions to permit droplet transfer at a much lower concentration of sucrose (approx. 1.5% w/v) that hardly affects WGE translation (Takahashi and Ogawa, 2020).
Our protocol with the optimized conditions allows us to prepare WGE-encapsulating SGUVs in which WGE translation takes place as efficiently as outside a liposome. The average successful formation rate of SGUVs is more than 90%, and the formed SGUVs are mostly stable for more than 60 min. The size of SGUVs can be controlled precisely (diameter = 1.6-4.4 mm) without impairing their formation or stability just by changing the volume of the droplet. No special equipment is required for the preparation or observation of SGUVs. Moreover, given that a WGE-based translation system enables the expression of a broad range of genes from various types of organisms (both eukaryotes and prokaryotes) as well as viruses (Madono et al., 2011), WGE-encapsulating SGUVs prepared using our protocol would be useful as a versatile research tool in cell-free synthetic biology (Hodgman and Jewett, 2012; Lentini et al., 2016; Kai and Schwille, 2019).
Materials and Reagents
Nuclease-free 2 ml tubes (WATSON, catalog number: 132-620C)
Nuclease-free PCR tubes (any brand)
Nuclease-free 1.7 ml tubes (any brand)
Nuclease-free 15 ml tubes (any brand)
Nuclease-free pipette tips (any brand)
Parafilm
384-well microplates (Greiner Bio-One, catalog number: 781801)
[For analyzing fluorescent protein] 96-well black plates (Corning, catalog number: 3915)
Nuclease-free water (Sigma-Aldrich, catalog number: W4502 or equivalent)
Linear double-stranded DNA encoding the protein of interest. The 5′ UTR sequence should be GAT TTA GGT GAC ACT ATAGAA CTC ACC TAT CTC CCC AAC ACC TAA TAA CAT TCA ATC ACT CTT TCC ACT AAC CAC CTA TCT ACA TCA CCA AGA TAT CAC TAG T (the SP6 promoter is underlined; a translational enhancer sequence named E01 is italicized; Kamura et al., 2005). Although any 3′ UTR sequence is permissible, its length should be more than 1,200 nt (Ogawa et al., 2014). Store at -20°C until use. See the instructions of the following kit for preparing the DNA.
SP6-Scribe Standard RNA IVT Kit (CellScript, catalog number: C-AS3106)
QIAquick Nucleotide Removal Kit (QIAGEN, catalog number: 28304) or RNeasy MinElute Cleanup Kit (QIAGEN, catalog number: 74204)
WEPRO1240 (CellFree Sciences, catalog number: CFS-WGE-1240). Divide into smaller volumes and store at -80°C
Nuclease-free sucrose (FUJIFILM Wako, catalog number: 198-13525)
Asolectin (FUJIFILM Wako, catalog number: 390-00271). Seal the container with parafilm and store at 4°C away from moisture
Mineral oil for molecular biology (Sigma-Aldrich, catalog number: M5904)
Creatine kinase (Roche, catalog number: 10127566001)
SUB-AMIX (CellFree Sciences, catalog number: CFS-SUB): a reaction buffer containing a mixture of substrates for translation such as amino acids and energy sources
Creatine kinase solution (see Recipes)
SUB-AMIX solution (see Recipes)
Translation solution (for twelve SGUVs with a volume of 2.5 μl) (see Recipes)
Equipment
Micropipettes (any brand)
Incubator (22°C, any brand)
Refrigerator (4°C, any brand)
Freezers (-80°C and -20°C, any brand)
Digital scale (any brand)
Vortex (any brand)
UV/vis microplate reader (Thermo Fisher Scientific, model: Multiskan GO) or an alternative instrument capable of RNA quantitation
[For analyzing fluorescent protein] Fluorescence plate reader (PerkinElmer, model: ARVO X2)
[For analyzing fluorescent protein] Blue-LED light source with an appropriate filter (OptoCode, model: LEDB-MBOXH or ATTO, model: Visirays-B)
Procedure
Note: The following protocol is for preparing twelve SGUVs. A schematic illustration of the procedure is shown in Figure 1.
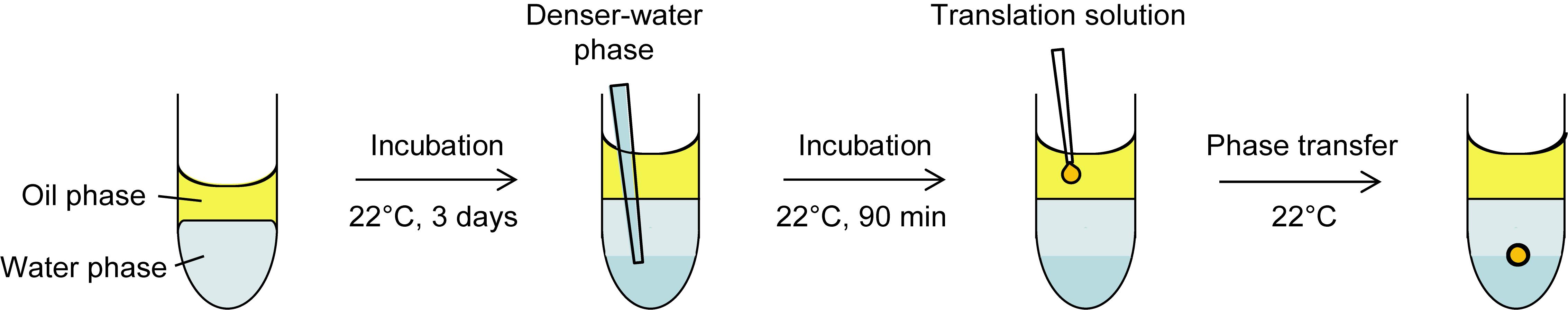
Figure 1. Schematic illustration of the procedure for preparing an SGUV. Refer to the following protocol for the detailed procedure. Adapted with permission from Takahashi and Ogawa (2020). Copyright 2020 American Chemical Society.
Preparation of the mRNA template
Transcribe the double-stranded DNA encoding the protein of interest (here, we use DNA encoding yellow fluorescent protein, YPet, as an example; Nguyen and Daugherty, 2005). Specifically, transcribe at 37°C for more than 2 h (preferably overnight) in a PCR tube containing 20 μl reaction solution using the SP6-Scribe Standard RNA IVT Kit according to the manufacturer’s protocol.
Digest the DNA by adding 1 μl DNase I included in the kit and incubating at 37°C for 15 min.
Purify the transcripts (i.e., mRNA) using the QIAquick Nucleotide Removal Kit or RNeasy MinElute Cleanup Kit according to each manufacturer’s protocol.
Note: Elute the mRNA from the QIAquick Nucleotide Removal Kit or RNeasy MinElute Cleanup Kit with 30 μl or 14 μl nuclease-free water, respectively.
Dilute 1 μl purified mRNA with 99 μl nuclease-free water and quantitate the diluted mRNA in a 384-well plate by measuring absorbance at 260 nm with the UV/vis microplate reader.
Note: Alternatively, other common spectroscopic or fluorescent approaches can be used to quantitate the mRNA.
Dilute the purified mRNA (from Step A3) with nuclease-free water to 1.5 μM.
Divide it into smaller volumes and store at -80°C until use.
Preparation of the oil phase, 0.5% w/v asolectin in mineral oil
Warm the container of asolectin at room temperature for 15 min.
Add 1.0 ml mineral oil to 20 mg asolectin in a 1.7-ml tube.
Seal the tube with parafilm.
Vortex the mixture to completely dissolve the asolectin to a 2% w/v solution.
Note: The volume change is negligible.
Mix 800 μl 2% w/v asolectin with 2.4 ml mineral oil in a 15-ml tube to prepare 3.2 ml 0.5% w/v asolectin.
Note: Use 0.5% w/v asolectin as soon as possible in Procedure D. Do not store.
Preparation of the water phase, 0.5× SUB-AMIX
Mix 25 μl 5× SUB-AMIX and 225 μl nuclease-free water in each of twelve 2-ml tubes to prepare 0.5× SUB-AMIX.
Layering the oil phase onto the water phase
Layer 250 μl 0.5% w/v asolectin (prepared in Procedure B) gently onto 250 μl 0.5× SUB-AMIX in each 2-ml tube (prepared in Procedure C).
Seal the tubes with parafilm.
Incubate the tubes at 22°C for 3 days in an incubator to form a lipid monolayer membrane at the oil/water interface.
Preparation of the denser water phase, 1.7× SUB-AMIX containing 3.5% w/v sucrose
Dissolve 5 g sucrose into about 7 ml nuclease-free water in a 15-ml tube.
Note: The volume changes significantly by dissolving sucrose in water.
Weigh the total volume and 1 ml sucrose solution to calculate the % w/v concentration using the following equation: % w/v = (5/weight of the total volume) × weight of 1 ml × 100.
Dilute the sucrose solution with nuclease-free water to 43.6% w/v.
Add 173 μl 43.6% w/v sucrose and 730 μl 5× SUB-AMIX to 1,232 μl nuclease-free water to prepare 2,135 μl 1.7× SUB-AMIX containing 3.5% w/v sucrose.
Layering the denser water phase under the water phase
Pipette 175 μl 1.7× SUB-AMIX containing 3.5% w/v sucrose (prepared in Procedure E) slowly to the bottom of each 2-ml tube (prepared in Procedure D); see Figure 1.
Notes:
To the greatest extent possible, do not disturb the lipid membrane at the oil/water interface.
The denser water phase is useful for preventing liposomes (SGUVs) from bursting by contacting the bottom of the tube and for balancing the osmotic pressure between the inside and outside of the liposomes after complete diffusion into the water phase.
Incubate the tubes at 22°C for 90 min in the same incubator.
SGUV formation
Prepare a cell-free translation solution containing approx. 1.5% w/v sucrose on ice (see Recipes).
Add an aliquot of the translation solution (2.0-45.0 μl) near the center of the oil phase in each 2-ml tube immediately after the 90-min incubation in Procedure F; see Figure 1.
Note: Although various sizes of SGUVs can be prepared within this range of volumes at a similar success rate, we recommend 2.5 μl for the first trial.
Incubate the tubes at 22°C in the same incubator and observe the phase transfer. Representative images of an incomplete 2.5-μl SGUV during the phase transfer and the resulting complete 2.5-μl SGUV are shown in Figure 2A and 2B, respectively.
Note: It takes several minutes for a droplet containing the translation solution to transfer through the oil/water interface and form an SGUV. The larger the volume, the faster the transfer becomes. The generated SGUVs are generally stable for more than 60 min.

Figure 2. Representative images of SGUV formation. A. An incomplete 2.5-μl SGUV during the phase transfer. B. The resulting complete 2.5-μl SGUV. The arrowheads show SGUVs. Scale bars: 2.5 mm. Adapted with permission from Takahashi and Ogawa (2020). Copyright 2020 American Chemical Society.
Detection of expressed protein in SGUVs (in the case of a fluorescent protein such as YPet)
Observe the fluorescence in SGUVs using the blue-LED light source with an appropriate filter. Figure 3 shows representative images of mRNA-encapsulating and mRNA-free SGUVs.
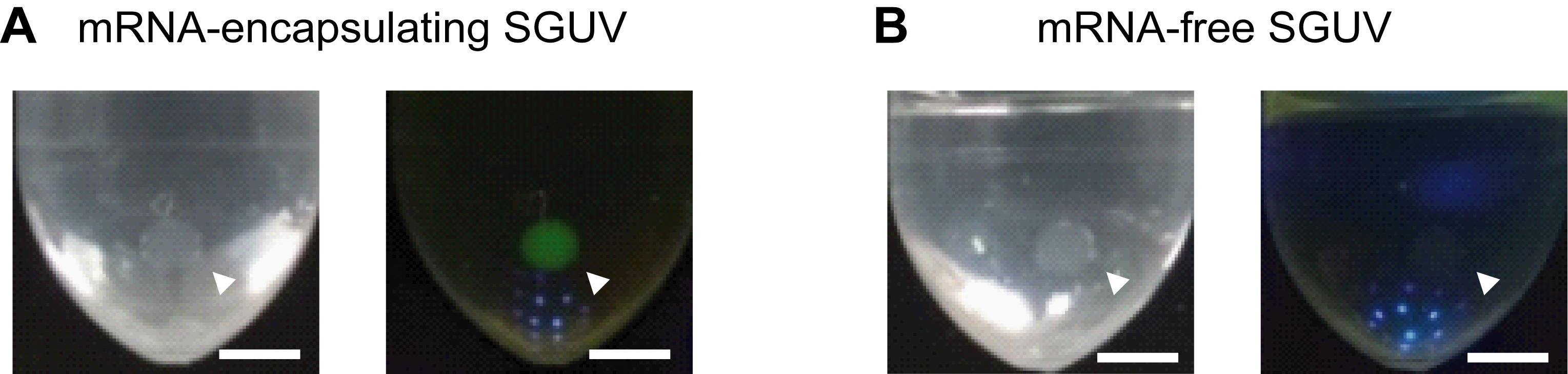
Figure 3. Representative images of SGUVs with and without mRNA encoding YPet in the interior. Visible light (left) and fluorescence (right) images of an mRNA-encapsulating SGUV (A) and an mRNA-free SGUV (B) 90 min after the addition of a droplet to the oil phase. The arrowheads show SGUVs. Scale bars: 2.5 mm. Adapted with permission from Takahashi and Ogawa (2020). Copyright 2020 American Chemical Society.To precisely measure the fluorescence intensity, use the following protocol.
Transfer 50 μl (or more for larger SGUVs) water phase containing the whole SGUV to a 1.7-ml tube.
Note: Use a pipette tip cut with clean scissors to be larger than that of the SGUVs.
Burst the SGUV by vortexing.
Transfer an aliquot (e.g., 45 μl) to a 96-well black plate.
Measure the fluorescence intensity using appropriate excitation/emission wavelengths (e.g., 485/535 nm) and exposure time (e.g., 1 s) in the fluorescence plate reader.
Recipes
Creatine kinase solution
Dissolve in nuclease-free water to make a 2 mg/ml solution, divide into smaller volumes, and store at -80°C.
SUB-AMIX solution
Since SUB-AMIX consists of a set of four reagents at 40× each concentration (S-1, S-2, S-3, S-4), prepare 8 ml 5× SUB-AMIX by adding 1 ml each S-1 through S-4 to 4 ml nuclease-free water, divide into smaller volumes, and store at -80°C.
Note: Do not mix the four reagents first.
Translation solution (for twelve SGUVs with a volume of 2.5 μl)
6.0 μl WEPRO1240; 0.6 μl 2 mg/ml creatine kinase; 6.0 μl 5× SUB-AMIX; 11.4 μl 3.8% w/v sucrose; 6.0 μl 1.5 μM mRNA (prepared in Procedure A) or nuclease-free water (for negative control)
Notes:
Prepare 3.8% w/v sucrose by diluting 43.6% w/v sucrose prepared in Procedure E3.
For a larger SGUV, increase each volume at an equal ratio.
Acknowledgments
This work was partially supported by JSPS KAKENHI Grant Number JP19K05697.
Competing interests
The authors claim no financial or non-financial competing interests.
References
- Elani, Y., Law, R. V., and Ces, O. (2015). Protein synthesis in artificial cells: using compartmentalisation for spatial organisation in vesicle bioreactors. Phys Chem Chem Phys 17(24): 15534-15537.
- Hamada, T., Miura, Y., Komatsu, Y., Kishimoto, Y., Vestergaard, M., and Takagi, M. (2008). Construction of asymmetric cell-sized lipid vesicles from lipid-coated water-in-oil microdroplets. J Phys Chem B 112(47): 14678-14681.
- Hodgman, C. E., and Jewett, M. C. (2012). Cell-free synthetic biology: thinking outside the cell. Metab Eng 14(3): 261-269.
- Kai, L., and Schwille, P. (2019). Cell-free protein synthesis and its perspectives for assembling cells from the bottom-up.Adv Biosys 3(6): 1800322.
- Kamura, N., Sawasaki, T., Kasahara, Y., Takai, K., and Endo, Y. (2005). Selection of 5′-untranslated sequences that enhance initiation of translation in a cell-free protein synthesis system from wheat embryos. Bioorg Med Chem Lett15(24): 5402-5406.
- Kubatta, E. A., and Rehage, H. (2009). Characterization of giant vesicles formed by phase transfer processes. Colloid Polym Sci 287(9): 1117-1122.
- Lentini, R., Yeh Martín, N., and Mansy, S. S. (2016). Communicating artificial cells. Curr Opin Chem Biol 34: 53-61.
- Madono, M., Sawasaki, T., Morishita, R., and Endo, Y. (2011). Wheat germ cell-free protein production system for post-genomic research. New Biotechnol 28(3): 211-217.
- Nguyen, A. W., and Daugherty, P. S. (2005). Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol 23(3), 355-360.
- Noireaux, V., and Libchaber, A. (2004). A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci USA 101(51): 17669-17674.
- Ogawa, A., Tabuchi, J., and Doi, Y. (2014). Identification of short untranslated regions that sufficiently enhance translation in high-quality wheat germ extract. Bioorg Med Chem Lett 24(16): 3724-3727.
- Pautot, S., Frisken, B. J., and Weitz, D. A. (2003). Production of unilamellar vesicles using an inverted emulsion. Langmuir 19(7): 2870-2879.
- Takahashi, H., and Ogawa, A. (2020). Preparation of a Millimeter-Sized Supergiant Liposome That Allows for Efficient, Eukaryotic Cell-Free Translation in the Interior by Spontaneous Emulsion Transfer. ACS Synth Biol 9(7): 1608-1614.
- Walde, P., Cosentino, K., Engel, H., and Stano, P. (2010). Giant vesicles: preparations and applications. ChemBioChem 11(7): 848-865.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Takahashi, H. and Ogawa, A. (2021). A Detailed Protocol for Preparing Millimeter-sized Supergiant Liposomes that Permit Efficient Eukaryotic Cell-free Translation in the Interior. Bio-protocol 11(12): e4054. DOI: 10.21769/BioProtoc.4054.
Category
Biological Engineering > Biomedical engineering
Biochemistry > RNA > mRNA translation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










