- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measuring Cytosolic Translocation of Mycobacterium marinum in RAW264.7 Macrophages with a CCF4-AM FRET Assay
Published: Vol 11, Iss 8, Apr 20, 2021 DOI: 10.21769/BioProtoc.3991 Views: 3611
Reviewed by: Jason NeidlemanMei Po ChanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In situ, Cell-free Protein Expression on Microarrays and Their Use for the Detection of Immune Responses
Katrin Hufnagel [...] Jörg D. Hoheisel
Feb 5, 2019 6545 Views

Transcervical Mouse Infections with Chlamydia trachomatis and Determination of Bacterial Burden
Karthika Rajeeve and Rajeeve Sivadasan
Feb 5, 2020 5915 Views
Abstract
The CCF4-AM Förster resonance energy transfer (FRET) assay is a sensitive approach to measure bacterial cytosolic translocation in live cells. The FRET pair hydroxycoumarin (donor) and fluorescein (acceptor) are linked by a CCF4-AM β-lactam ring, the substrate of β-lactamase. The exogenously added, neutral charged-FRET reagent can diffuse across the membrane and stay in the cytosol only once it is charged in the cytosol. When bacteria translocate from subcellular organelles (e.g., phagosomes) to the cytosol, the bacteria-associated β-lactamase cleaves the β-lactam ring, resulting in loss of FRET signal. Here we describe the fluorometer-based approach optimized for direct measurement of cytosolic translocation as a result of the EsxAB complex of Mycobacterium marinum in RAW264.7 cells.
Keywords: CCF4-AMBackground
The development of a β-lactamase reporter assay gave rise to the subsequent Förster resonance energy transfer (FRET) assay (Zlokarnik et al., 1998; Cavrois et al., 2002). Two fluorescence dyes, hydroxycoumarin (donor) and fluorescein (acceptor), are linked via a CCF4-AM (CCF2-AM in earlier reports) β-lactam ring. When excited at 405 nm, hydroxycoumarin emits at 450 nm (blue), which subsequently excites fluorescein to emit at 527 nm (green). Cleavage of the β-lactam ring by bacteria-associated β-lactamase results in loss of FRET and direct emission of hydroxycoumarin (blue) (Jones and Padilla-Parra, 2016). The CCF4-AM FRET reporter assay is versatile and has been been used for a variety of different applications, such as tracking molecular fusions (Jones and Padilla-Parra, 2016; Shaikh, et al., 2019), tracking delivery of biological cargos to the cytosol (Stone et al., 2018), and monitoring translocation of bacteria within cells (Charpentier and Oswald, 2004). Since Mycobacterium marinum also possesses β-lactamase (Flores et al., 2005), Simeone and colleagues exploited the FRET assay to track cytosolic translocation of Mycobacterium tuberculosis in THP-1 cells (Simeone et al., 2012). Recently, we applied this method for investigation of the role of EsxAB in cytosolic translocation of Mycobacterium marinum (Acosta et al., 2014; Zhang et al., 2016; Aguilera et al., 2020). RAW264.7 cells were infected with M. marinum and the results were obtained under a fluorometer, providing a quick, quantitative method for analysis of results. Furthermore, as Mycobacterium marinum is a BSL-2 pathogen and contains similarities in its ESX-1 locus (Tobin and Ramakrishnan, 2008), it provides a safe, direct way to investigate the EsxAB complex. Here we describe our procedure for direct monitoring of cytosolic translocation of Mycobacterium marinum in RAW264.7 cells via the CCF4-AM FRET assay using a fluorometer.
Materials and Reagents
- Inoculation loop (BD, catalog number: 220216)
- Cell Scraper (Fisherbrand, catalog number: 08-100-241)
- T-75 Flask (Thermo Scientific, catalog number: 12-566-440)
- 50 ml Centrifuge tube (Corning, catalog number: 352098)
- 6-well plate (Corning, catalog number: 3516)
- 10 ml syringe (Fisherbrand, catalog number: 14-955-459)
- 22 gauge needle (BD, catalog number: 305900)
- 27 gauge needle (BD, catalog number: 305109)
- 5 µm filter (Millex, catalog number: SLSV025LS)
- Pasteur pipette (Fisherbrand, catalog number: 13-678-6b)
- Aluminum Foil (Fisherbrand, catalog number: 01-213-102)
- Thin Wall Cuvette (Fireflysci, catalog number: 30FL)
- 435 nm long-pass filter (Andover Corporation, catalog number: 435FG03-50S)
- 7H9 media (Difco, catalog number: 271310)
- OADC Enrichment (BD, catalog number: 211886)
- Tween-80 (Sigma-Aldrich, catalog number: P1754)
- Kanamycin Monosulfate (Gold Bio, catalog number: K-120-5)
- DMEM media (Gibco, catalog number: 10566-016)
- Fetal Bovine Serum (Gibco, catalog number: A4766801)
- Penicillin and Streptomycin (Gibco, catalog number: 15070063)
- Phosphate buffered saline (Sigma-Aldrich, catalog number: P3813)
- CCF4-AM FRET Loading Solutions (Life Technologies, catalog number: K1085)
Note: This kit contains Solution A, Solution B, and Solution C as described in the procedure. - DMSO (Sigma-Aldrich, catalog number: D8418)
- 4% Paraformaldehyde
- NaCl (Fisherbrand, catalog number: 18-606-420)
- MgCl (Fisherbrand, catalog number: 18-604-047)
- CaCl2 (Fisherbrand, catalog number: AA4428030)
- KCl (Spectrum Chemical, catalog number: 18-605-507)
- Glucose (Sigma-Aldrich, catalog number: G7021)
- 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid (HEPES)) (Fisher Bioreagents, catalog number: BP310)
- Tris (Fisher Bioreagents, catalog number: BP152-500)
- EM Media (see Recipes)
Equipment
- Biosafety Cabinet (Labconco)
- Vortex (VWR, catalog number: 58816-121)
- Hemocytometer (Hausser Scientific, catalog number: S17040)
- Inverted Light Microscope (VWR, catalog number: Vistavision)
- Cell Imaging System (Life Technologies, catalog number: 4471136)
- Spectrophotometer (Amersham Biosciences, model: Ultrospec 10)
- Table Top Centrifuge (Eppendorf, catalog number: 5810R)
- CO2 Incubator (VWR, catalog number: 3078)
- Fluorometer (ISS, catalog number: K2)
- Incubator Shaker (VWR, catalog number: 1585)
- Hot Plate/Stirrer (ThermoFisher Scientific, catalog number: SP131325)
- pH meter (Mettler Toledo, catalog number: 30266626)
Procedure
- Growth of M. Marinum bacterial culture
- Approximately 5 d before the infection assay, prepare the M. Marinum culture for infection.
- Inoculate M. marinum into 50 ml of 7H9 media supplemented with 10% OADC enrichment and proper antibiotics as needed, we routinely use 30 mg/ml Kanamycin.
- This bacterial culture will be used to start a new bacterial culture and passaged to reduce cell clumping.
- If the bacterial culture contains too many clumped cells, then add Tween-80 at a concentration of 0.05% (v/v) to the 7H9 media.
- Allow the bacterial culture to grow at 30 °C while shaking at 150 rpm until it reaches approximately OD600 1.2.
- Once the bacterial culture has grown past mid log phase, transfer 4 ml of culture into 46 ml of fresh 7H9 media supplemented with 10% OADC.
- Incubate at 30 °C while shaking at 150 rpm; allow the bacterial culture to grow into mid log phase once more, approximately 2 d.
- Culturing of RAW264.7 mouse macrophage cells
- Approximately 2-3 d before infection, grow RAW264.7 cells in a T-75 flask with 10 ml of DMEM media containing 10% Fetal Bovine Serum (FBS) with penicillin and streptomycin at 100 unit/ml.
- Allow the cells to grow to 80-90% confluence at 37 °C with 5% CO2.
- The day before the infection, use a cell scraper to lift all of the cells (at 80-90% conflucence) from the bottom of the TC flask.
- Transfer the cells and media into a 50 ml centrifuge tube.
- Centrifuge at 160 × g for 10 min at RT.
- Remove supernatant media and discard.
- Resuspend the pellet in 10 ml of DMEM media containing 10% FBS with penicillin and streptomycin.
- Count the cells using whichever method you prefer. Record the approximate number of cells in the sample.
- Fill a 6-well plate with 2 ml of media per well, then add RAW264.7 cells to seed a total of 2.5 × 106 cells/well.
- Incubate the cells overnight at 37 °C with 5% CO2.
- Bacterial single-cell preparation
- Pellet down the bacterial culture from Part A by centrifuging at 3,220 × g for 15 min.
- Remove the supernatant and resuspend the bacteria in 50 ml of PBS.
- Wash the cells 2 more times for a total of 3 washes with PBS.
- After the third wash, remove the supernatant and resuspend the pellet in 10 ml of PBS.
- Aspirate the bacteria in PBS using a syringe and pass through a 22-gauge needle.
Note: This step is optional but will facilitate passing the culture through a filter. - Aspirate the bacteria with a syringe and pass through a 27-gauge needle.
- Aspirate the bacteria and pass through a 5 µm filter.
- Measure the density of the bacterial culture with OD600.
- Calculate the volume of bacterial cells needed to add to your macrophages depending on the multiplicity of infection (MOI).
- If needed, concentrate the bacterial cells by pelleting and resuspending them into a smaller volume.
- Infection of macrophages
- Check RAW264.7 cells under a microscope; they should be at 60% confluence.
- Aspirate DMEM media and replace with EM medium.
- Infect macrophages with Mycobacterium marinum at 10 MOI, or at the desired MOI.
- Incubate for 2 h at 30 °C with 5% CO2.
- After incubation, aspirate the media.
- Gently wash with 3 ml PBS and agitate the plate.
- Remove the PBS and wash 2 more times to remove extracellular bacteria.
- Add 2.5 ml of pre-warmed EM media with 10% FBS and incubate for 48 h.
- CCF4-AM FRET Assay
- Load RAW264.7 cells with CCF4-AM loading solutions by following the manufacturer’s recommendation:
- The uninfected RAW264.7 cells loaded with CCF4-AM serve as a negative control.
- The RAW264.7 cells without CCF4-AM serve as another negative control.
- A Chiense hamster ovary (CHO) cell line that expresses β-lactamase (CHO-β-Lac) is used as a positive control for cleavage of CCF4-AM (Zhang et al., 2016).
- Solution B (100 mg/ml Pluronic-F127 surfactant in DMSO and 0.1% acetic acid) and Solution C (24% w/w PEG 400, 18% TR-40 v/v in water) are prepared by manufacturer.
- Add 182 µl of DMSO to dissolve 200 µg of CCF4-AM, gently pipet up and down.
Note: This will be solution A, as specified by manufacturer. - Decant the solution into 20 µl aliquots.
Notes:- If you wish to store, desiccate under vacuum and store at -20 °C protected from light.
- If vacuum is not available, the solution may be stored at -20 °C in the dark. This solution is stable for up to three months.
- Add 3 µl of Solution A to 30 µl of Solution B and vortex.
- Add 467 µl of Solution C and vortex to produce 6× CCF4-AM substrate loading solution.
Note: This solution is stable for 12 h at RT. - Add 500 µl of 6× CCF4-AM loading solution to 100 µl of the cell culture to make 1× CCF4-AM loading solution.
- Incubate at room temperature for 2 h while protected from light.
Note: If cells are disturbed, the 2-h incubation will give them enough time to settle to the bottom of the plate. - Aspirate the media and CCF4-AM substrate.
- Resuspend the cells with 3 ml PBS and agitate the plate gently.
- Remove the PBS and wash 2 more times with PBS for a total of 3 washes.
- Fix the cells with fresh 4% paraformaldehyde (PFA) for 30 min at room temperature (RT) while protected from light.
- Wash once more with PBS and at this stage the cells are ready for measurement.
- Load RAW264.7 cells with CCF4-AM loading solutions by following the manufacturer’s recommendation:
- Measurement using a fluorometer
- Gently resuspend the cells in PBS and transfer them to a thin walled UV cuvette.
- The fluorometer program should be set up to excite at 405 nm and record emissions between 430 nm to 600 nm to obtain the curves containing both blue and green peaks (Figure 1).
Note: In the fluorometer, two polarizers are placed vertical to each other on the excitation beam path and a 435 nm long-pass filter is placed on the emssition beam path to reduce the background and scattered light.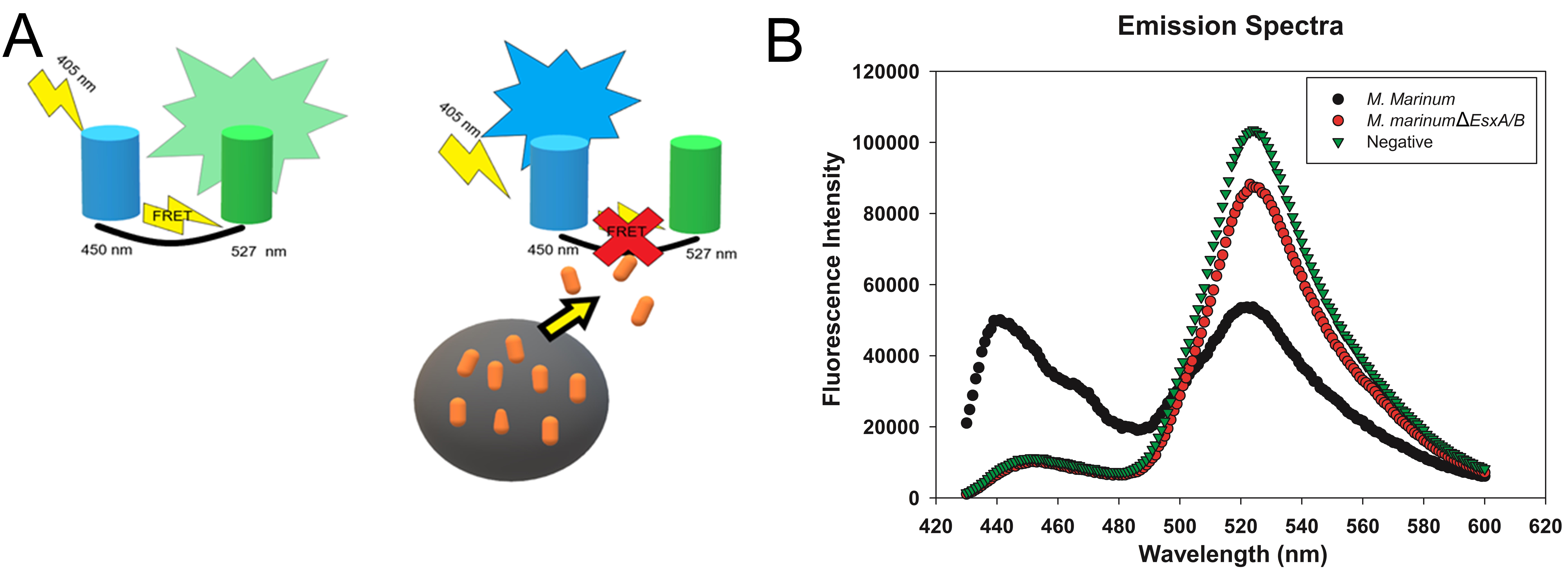
Figure 1. The emission spectra of the samples were recorded from FRET measurement. A. After excitation at 405 nm, the hydroxycoumarin will transfer energy to fluorescein resulting in a green emission at 527 nm. Bacteria containing β-lactamase will escape the phagosome and come into contact with the FRET dye in the cytosol, resulting in the loss of FRET and a blue emission at 450 nm. B. The first peak is the emission of hydroxycoumarin (blue) and the second peak is the emission of fluorescein (green). - The intensitiy at 450 nm (blue), I1, and the intensity at 527 nm, I2, are measured for 20 iterations. The Blue/Green ratio is calculated as I1/I2 (Figure 2).
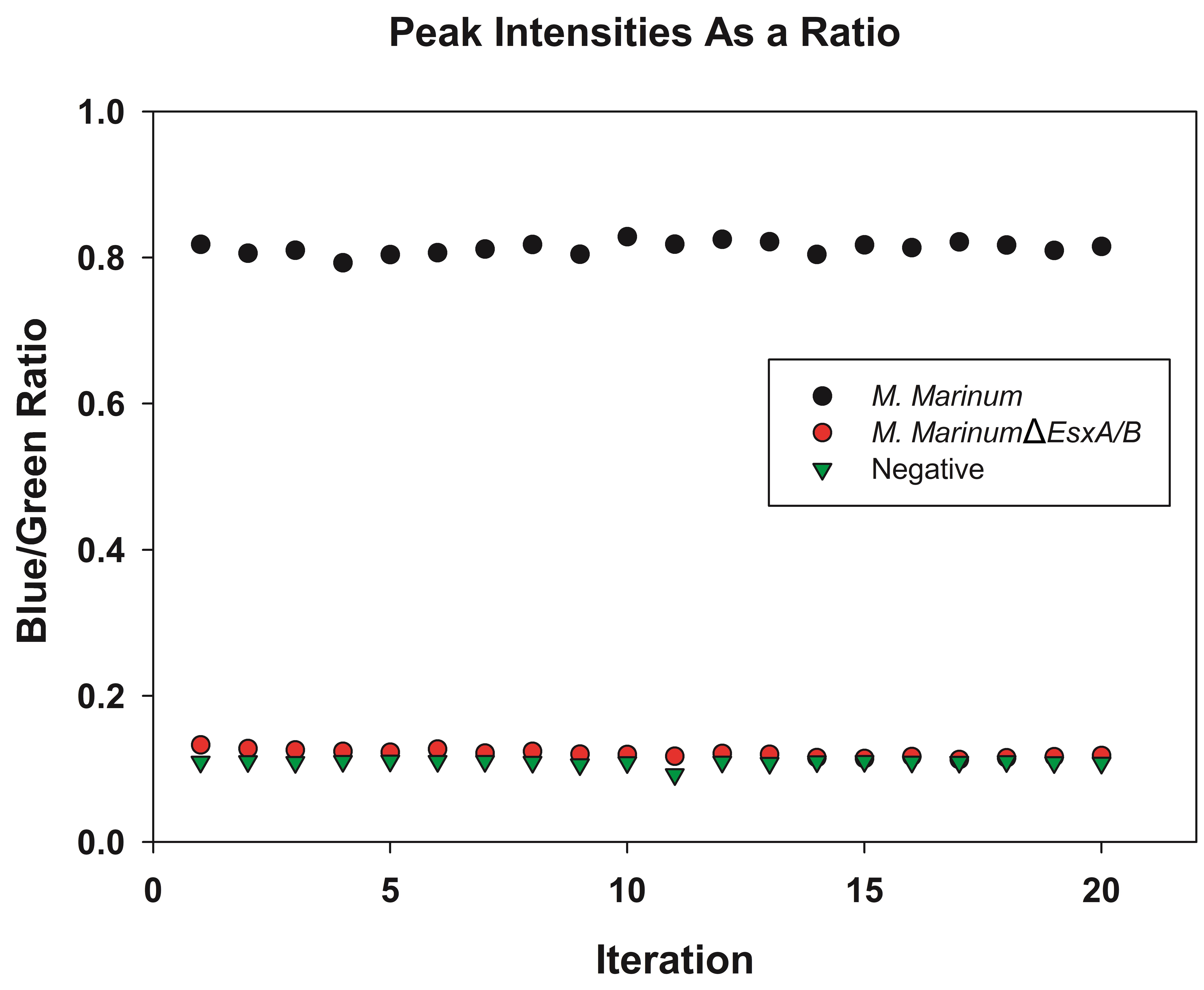
Figure 2. The blue/green ratio for sample data is calculated by division of I1 by I2. The intensity at 450 nm (blue) and 527 nm (green) are measured for 20 iterations. The Blue/Green ratio is calculated. A higher ratio, as observed for Mycobacterium marinum of ~0.8 represents a good change of signal, indicating cytosolic translocation. Lower ratios (< 0.2) are obtained by bacteria unable to escape the phagosome or the negative control.
Data analysis
- The Blue/Green ratios are averaged for 20 iterations from 3 duplicates, a total of 60 replicates.
- The average Blue/Green ratio is then plotted as a bar graph with error bars representing standard deviation and tested for statistical significance (Figure 3).
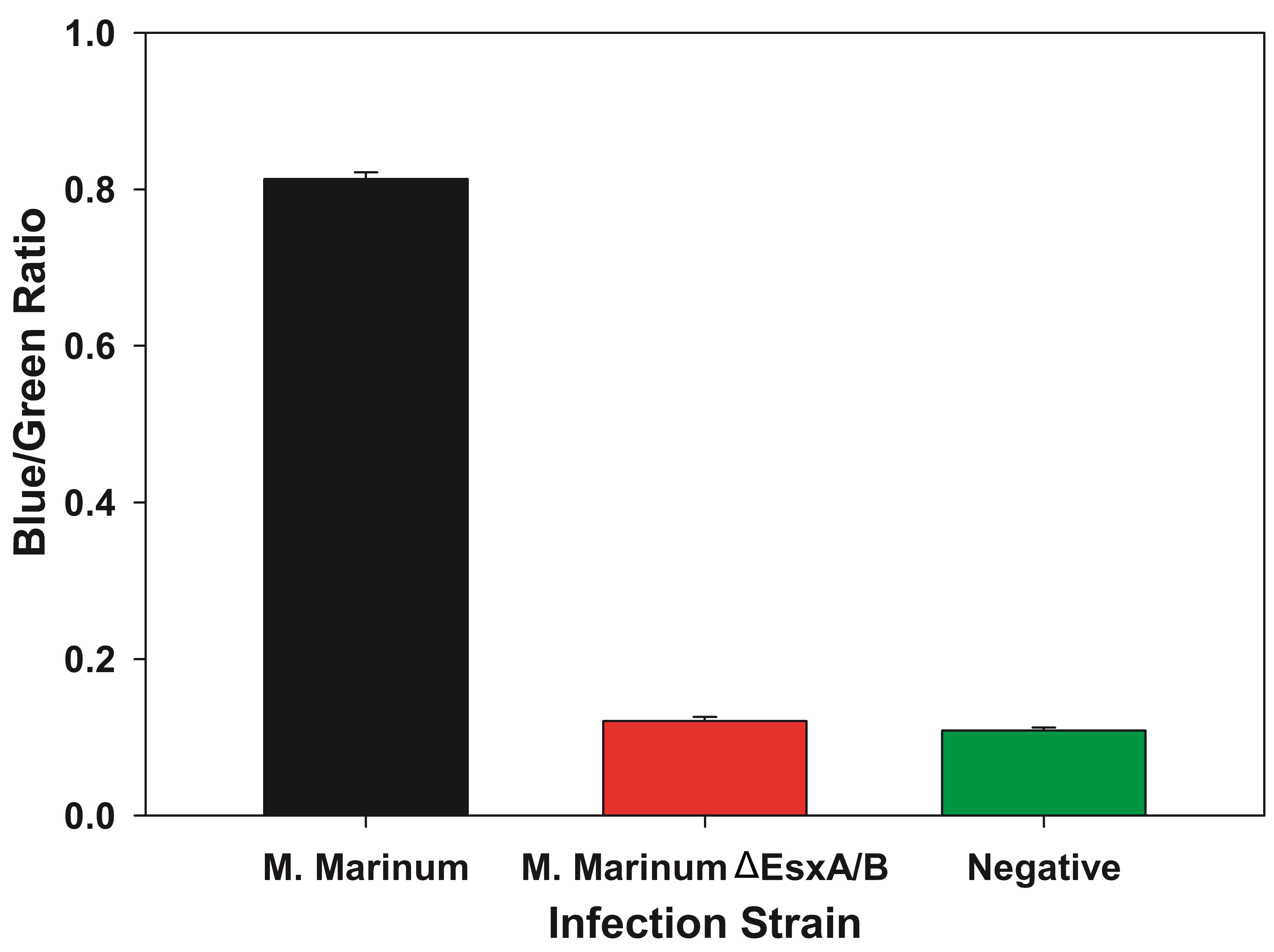
Figure 3. Sample data as bar graph with Blue/Green ratio for different strains. By plotting the data in a bar graph, the loss of FRET between samples can be seen. In this scenario, the wild type M. marinum escapes the phagosome and interacts with the FRET dye resulting in a higher Blue/Green ratio. The M. marinumΔEsxA/B is unable to escape the phagosome and does not interact with the FRET dye resulting in no change of color from green to blue. - The values are tested for normality using the Shapiro-Wilk test.
- If the data fails to differ from normal, it is tested for significance with a One-way ANOVA, otherwise a Tukey-Kramer test is performed to test for statistical significance.
Recipes
- EM Media (1 L)
- In a beaker add 7.02 g of NaCl, 0.52 g of KCl, 0.26 g of CaCl2, 0.076 g of MgCl, 0.9 g glucose and 6.5 of HEPES to water
- Bring the pH to 7.3
- Bring water up to 1 L
Acknowledgments
The study is supported by the grants from NIGMS (SC1GM095475 to J. Sun), National Center for Research Resources (5G12RR008124) and National Institute on Minority Health and Health Disparities (G12MD007592). Javier Aguilera was supported by National Institutes of Health Grant (R25GM069621 to R. Aguilera), via the RISE program for graduate students. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interest
The authors have no competing interests to report.
References
- Acosta, Y., Zhang, Q., Rahaman, A., Ouellet, H., Xiao, C., Sun, J., and Li, C. (2014). Imaging cytosolic translocation of Mycobacteria with two-photon fluorescence resonance energy transfer microscopy. Biomed Opt Express 5(11): 3990-4001.
- Aguilera, J., Karki, C. B., Li, L., Reyes, S. V., Estevao, I., Grajeda, B. I., Qi Zhang, Arico, C. D., Ouellet, H. and Sun, J. (2020). Nα-Acetylation of the virulence factor EsxA is required for mycobacterial cytosolic translocation and virulence. J Biol Chem 295(17): 5785-5794.
- Cavrois, M., de Noronha, C., and Greene, W. C. (2002). A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol 20(11): 1151-1154.
- Charpentier, X. and Oswald, E. (2004). Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 β-lactamase as a new fluorescence-based reporter. J Bacteriol 186(16): 5486-5495.
- Flores, A. R., Parsons, L. M., and Pavelka Jr, M. S. (2005). Genetic analysis of the β-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to β-lactam antibiotics. Microbiology 151(2): 521-532.
- Jones, D. M., and Padilla-Parra, S. (2016). The β-lactamase assay: Harnessing a FRET biosensor to analyse viral fusion mechanisms. Sensors 16(7): 950.
- Simeone, R., Bobard, A., Lippmann, J., Bitter, W., Majlessi, L., Brosch, R., and Enninga, J. (2012). Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog 8(2): e1002507.
- Shaikh, F., Zhao, Y., Alvarez, L., Iliopoulou, M., Lohans, C., Schofield, C. J., Parra, S. P., Siu, S. W. I., Fry, E. E. and Ren, J. S. (2019). Structure-Based in silico screening identifies a potent ebolavirus inhibitor from a traditional chinese medicine library. J Med Chem 62(6): 2928-2937.
- Stone, S. R., Heinrich, T., Juraja, S. M., Satiaputra, J. N., Hall, C. M., Anastasas, M., Mills ,D. M., Chamberlain, C. A., Winslow, S. and Priebatsch, K. (2018). β-Lactamase tools for establishing cell internalization and cytosolic delivery of cell penetrating peptides. Biomolecules 8(3): 51.
- Tobin, D. M., and Ramakrishnan, L. (2008). Comparative pathogenesis of Mycobacteriummarinum and Mycobacterium tuberculosis. Cell Microbiol 10(5): 1027-1039.
- Zhang, Q., Wang, D., Jiang, G., Liu, W., Deng, Q., Li, X., Qian, W., Ouellet, H. and Sun, J. (2016). EsxA membrane-permeabilizing activity plays a key role in mycobacterial cytosolic translocation and virulence: effects of single-residue mutations at glutamine 5. Sci Rep 6: 32618.
- Zlokarnik, G., Negulescu, P. A., Knapp, T. E., Mere, L., Burres, N., Feng, L., Whitney, M., Roemer, K. and Tsien, R. Y. (1998). Quantitation of transcription and clonal selection of single living cells with β-lactamase as reporter. Science 279(5347): 84-88.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Aguilera, J. and Sun, J. (2021). Measuring Cytosolic Translocation of Mycobacterium marinum in RAW264.7 Macrophages with a CCF4-AM FRET Assay. Bio-protocol 11(8): e3991. DOI: 10.21769/BioProtoc.3991.
- Aguilera, J., Karki, C. B., Li, L., Reyes, S. V., Estevao, I., Grajeda, B. I., Qi Zhang, Arico, C. D., Ouellet, H. and Sun, J. (2020). Nα-Acetylation of the virulence factor EsxA is required for mycobacterial cytosolic translocation and virulence. J Biol Chem 295(17): 5785-5794.
Category
Microbiology > Microbe-host interactions > Bacterium
Cell Biology > Cell-based analysis > Bacterial infection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











