- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Pulse–chase EdU Method for Detection of Cell Division Orientation in Arabidopsis and Juncus prismatocarpus Leaf Primordia
Published: Vol 11, Iss 1, Jan 5, 2021 DOI: 10.21769/BioProtoc.3882 Views: 4877
Reviewed by: Takumi HigakiAleksandr GavrinAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Direct Plant Regeneration From Immature Male Inflorescence of Banana (Musa spp.)
Pradeep Chand Deo
Oct 20, 2025 1438 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views
Abstract
In plants, the morphological diversity of leaves is largely determined by cell division, especially cell division orientation. Whereas cell division itself is easily monitored, the detection and quantification of cell division orientation are difficult. The few existing methods for detection and quantification of cell division orientation are either inefficient or laborious. Here, we describe a pulse-chase strategy using a 5-ethynyl-2’-deoxyuridine (EdU) labeling assay. Plant tissues are first incubated with EdU for a short period (pulse), followed by a long incubation without EdU (chase). Using this method, the positions of daughter cells are easily detected and can be used to quantify cell division orientation. Our protocol is rapid and very efficient for quantitative analysis of cell division orientation, and can be applied to both model and non-model plant species.
Graphic abstract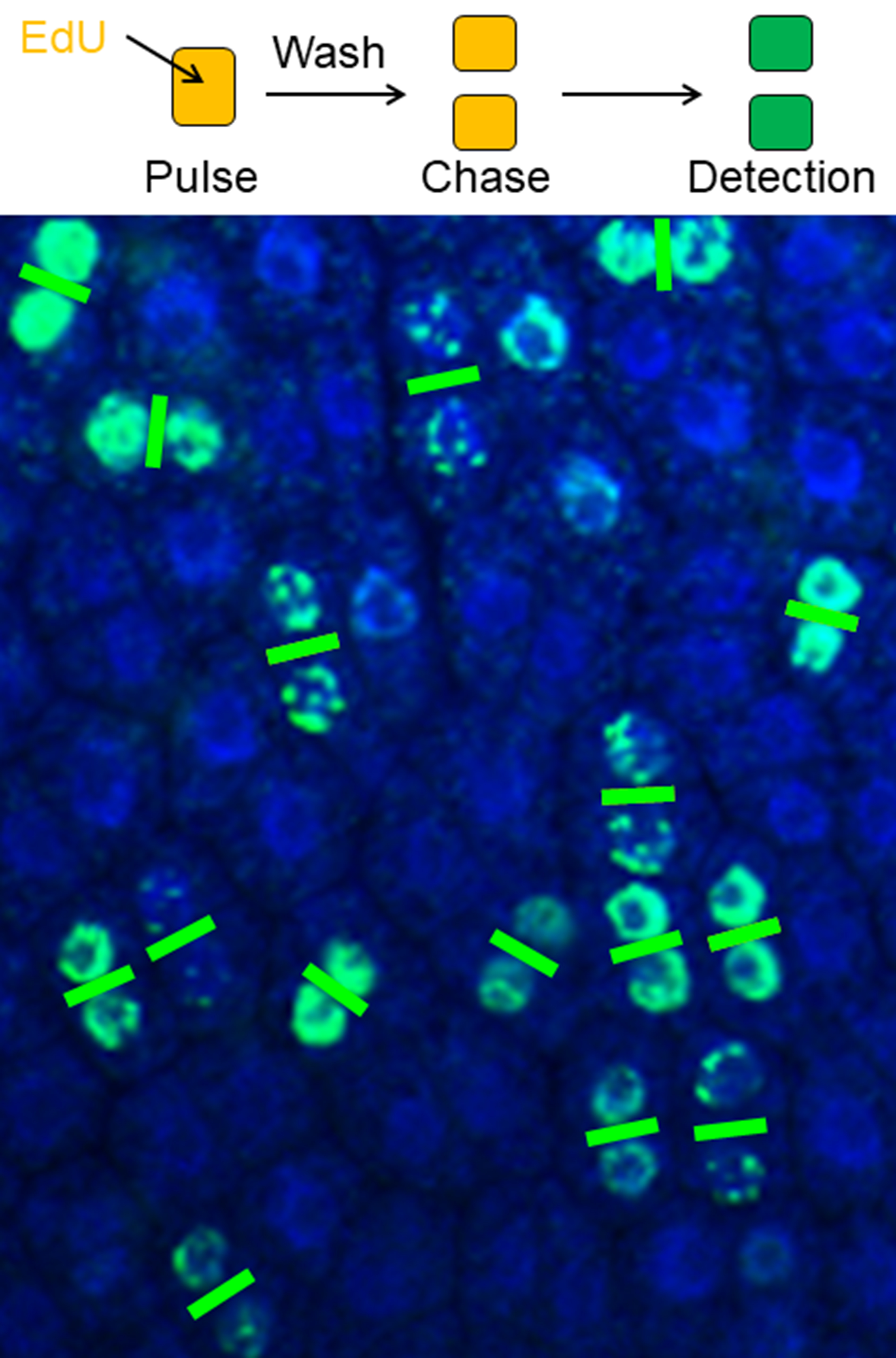
Plant cell division pairs clearly visualized by a pulse-chase EdU method
Background
Plant cells are attached to each other by cell walls and cannot migrate. Therefore, organized, oriented cell division during the early stages of leaf development largely determines the shapes of mature leaves. Thus far, no method for efficient and rapid detection and quantification of cell division orientation has been reported. Existing methods include visualization of daughter nuclei (in telophase) using a pCYCB1;1::GUS (β-glucuronidase) reporter line (Horiguchi et al., 2011) or visualization of spindle equators (in metaphase) using 4’,6-diamidino-2-phenylindole (DAPI) staining (Fukushima et al., 2015). However, these methods require considerable processing time and effort to identify cells from a particular phase. Recently, 5-ethynyl-2’-deoxyuridine (EdU), a thymidine analog, has become the agent of choice for monitoring cell division activity (Kotogany et al., 2010; Ichihashi et al., 2011; Nakayama et al., 2012, 2014 and 2015; Furuya et al., 2018; Koga et al., 2020). By combining a pulse-chase strategy and EdU staining, we developed a method that can rapidly detect large numbers of cell pairs (two daughter cells after a cell division event) over a short period of time (Yin and Tsukaya, 2016). In brief, plant tissues are first incubated with EdU for a short period (pulse), during which EdU is incorporated in cells in S phase. The pulse period is followed by a long incubation without EdU (chase), allowing EdU-carrying cells to proceed through the subsequent G2 and M phases. By optimization of the lengths of pulse and chase periods, pairs of daughter cells can be detected, because they contain identical amounts of EdU. Cell division orientation can be deduced according to the positions of the two daughter cells. Using this method, we showed that cell division orientation is locally non-stochastic in leaf primordia of Arabidopsis thaliana (Arabidopsis), a model plant species for which leaf morphogenesis has been extensively studied (Yin and Tsukaya, 2016). Furthermore, we combined this method with conventional paraffin sectioning and analyzed cell division orientation in leaves with more complex structures such as those of Juncus prismatocarpus, a non-model plant species (Yin and Tsukaya, 2019).
Materials and Reagents
1.5- or 5-ml tube
APS-coated glass slide (Matsunami, catalog number: SAPS )
Coverslip (Matsunami, catalog number: C025601 )
Razor blade
Aluminum foil
Gloves
Arabidopsis seed
J. prismatocarpus seed
10x phosphate buffered saline (PBS) (pH 7.4) (Nacalai Tesque, catalog number: 27575-31 ) (store at room temperature)
0.5% (v/v) Triton X-100 in 1x PBS (pH 7.4) (store at room temperature)
Click-iTTM EdU Cell Proliferation Kit for Imaging (Thermo Fisher Scientific, catalog number: C10337 ) (store at 4 °C and protect unopened components from light; store stock and working solutions following the manufacturer’s specifications)
DAPI (Thermo Fisher Scientific, catalog number: D1306 ) (store at 4 °C and protect from light)
Höechst 33342 (Component G of 11)
Ethanol (Wako, catalog number: 057-00451 )
Ethanol series (30%, 50%, 70%, 90%, and 100%) (store at room temperature)
Xylene (Wako, catalog number: 244-00081) (store at room temperature, preferably in a fume hood)
(Optional) Histo-Clear (National Diagnostics, catalog number: HS-200 )
Paraffin (McCormick Scientific, catalog number: 39502004 ) (store at room temperature and avoid moisture)
Distilled water (dH2O)
Sucrose (Nacalai Tesque, catalog number: 30403-55 )
Vitamin B5 (Wako ASB-00022727-001 )
Agar (Wako, catalog number: 016-11875 )
1 M KOH
Formaldehyde (Wako, catalog number: 064-00406 )
Glacial acetic acid (Wako, catalog number: 017-00251 )
Murashige and Skoog (MS) plant salt mixture (Wako, catalog number: 392-00591 )
MS plates and liquid MS medium (see Recipes) (store at 4°C)
EdU detection cocktail (see Recipes)
FAA (see Recipes) (store at room temperature)
Equipment
Forceps
Shaker (AS ONE, model: SRR-2 )
Confocal laser scanning microscope (Zeiss, model: LSM 780 )
Incubator (Taitec, model: HB-80 )
Paraffin mold (multiple)
Rotary microtome (Leica, model: Microm HM 355S )
Slide warmer (Tissue Tek, model: PS-52S )
Slide holder
Glass tank (multiple)
Software
Zen 2012 blue edition
Procedure
Arabidopsis leaf primordia
Germinate surface-sterilized Arabidopsis seeds on MS plates. At six days after sowing (DAS) at 22 °C under continuous light (70 μmol·m-2·sec-1 from white florescent lamps), select healthy seedlings and cut the root and part of the hypocotyl (Figure 1) using a razor blade.

Figure 1. Uncut (left) and cut (right) Arabidopsis samples. Scale bar = 1 mm.Transfer samples to a 1.5- or 5-ml tube approximately half-filled with liquid MS medium containing 10 μM EdU. Incubate for 45 min under continuous light at room temperature.
Transfer samples to a new tube filled with liquid MS medium without EdU. Gently shake [≤ 50 revolutions per minute (rpm)] for 5 min, three times.
Incubate samples in liquid MS medium for 6 h 45 min under continuous light at room temperature. The total chase time is 7 h, including the wash in Step A3.
Transfer samples to a new tube filled with FAA to fix the samples overnight at 4 °C. Degas by vacuum infiltration a few times until the samples sink. Alternatively, briefly centrifuge (130 x g, 5 s) to enhance fixation.
(Optional) Incubate samples in 90% acetone on ice for 10 min.
Shake (100 rpm) samples in 0.5% (v/v) Triton X-100 in 1x PBS (pH 7.4) for 5 min, two times.
Shake (100 rpm) samples in 1x PBS (pH 7.4) for 5 min, two times.
Prepare EdU detection cocktail (see Recipes) in a new tube. Transfer samples to it and incubate for 30 min in the dark.
Transfer samples to a new tube filled with 1x PBS (pH 7.4) and shake (100 rpm) for 5 min, three times.
Transfer samples to a new tube filled with 1x PBS (pH 7.4) containing 1 μg/ml DAPI and incubate for 30 min in the dark.
Shake (100 rpm) samples in 1x PBS (pH 7.4) for 20 min, two times and a third time overnight.
Mount samples with 1x PBS (pH 7.4) on a slide and observe the first pair of leaf primordia using a confocal laser scanning microscope equipped with a 40x oil immersion objective. Use a 488 nm laser to excite Alexa 488 (now covalently bonded to EdU) and a 490-535 nm emission spectrum to detect Alexa 488. Use a 405 nm laser to excite DAPI and a 420-470 nm emission spectrum to detect DAPI.
Store samples in 1x PBS (pH 7.4) in the dark at 4 °C for up to 6 months.
Juncus prismatocarpus leaf primordia
Germinate surface-sterilized J. prismatocarpus seeds on MS plates. At 28 DAS at 22 °C under continuous light (70 μmol·m-2·sec-1 from white florescent lamps), select healthy seedlings and cut the upper parts of leaves and elongated roots (Figure 2) using a razor blade.

Figure 2. Uncut (left) and cut (right) Juncus prismatocarpus samples. Scale bar = 5 mm.Transfer samples to a 1.5- or 5-ml tube approximately half-filled with liquid MS medium containing 10 μM EdU. Incubate for 2 h under continuous light at room temperature.
Transfer samples to a new tube filled with liquid MS medium without EdU. Gently shake (≤ 50 rpm) for 5 min, three times.
Incubate samples in liquid MS medium for 13 h 15 min under continuous light at room temperature. The total chase time is 13.5 h, including the wash in Step B3.
Transfer samples to a new tube filled with FAA to fix samples overnight at 4 °C. Degas by vacuum infiltration for a few times until the samples sink.
Dehydrate samples through an increasing series of ethanol concentrations, including 50%, 70%, 90%, and three changes of 100% ethanol. Incubate samples in each solution for 1 h.
Replace ethanol with xylene through an increasing series of xylene/ethanol concentrations, including 25%, 50%, 75%, and three changes of 100% xylene. Incubate samples in each solution for 30 min.
Infiltrate samples with paraffin through an increasing series of paraffin/xylene concentrations at 60 °C, including 25%, 50%, 75%, and 100% paraffin. Incubate samples in 25% paraffin/xylene overnight, in 50% and 75% paraffin/xylene for 4 h on the second day, and in 100% paraffin during the second overnight.
Incubate samples in paraffin for three additional days at 60 °C. Replace the 100% paraffin three times per day at a minimum interval of 4 h.
Embed samples in the desired orientation in a mold by solidifying the paraffin at room temperature or placing the mold on ice.
Section samples (8 μm) using a rotary microtome. Add dH2O on top of a glass slide until dH2O covers its entire surface. Mount serial sections on dH2O. Heat the slide on a slide warmer at 42 °C overnight.
Place slides (now containing sectioned samples) on a slide holder. Clear the paraffin by allowing the slide holder to pass through glass tanks containing the following in order: 100% xylene for 10 min, two times; 50% xylene/ethanol for 5 min; and 100% ethanol for 5 min.
Rehydrate samples by allowing the slide holder to pass through glass tanks containing the following in order: 90%, 70%, 50%, and 30% ethanol for 1 min each; dH2O for 5 min, two times; and 1x PBS (pH 7.4) for 5 min, two times.
Briefly remove the liquid from the slides and apply 300 μl freshly prepared EdU detection cocktail to each slide using a pipette. Gently cover the slide with a coverslip and incubate for 30 min in the dark.
Briefly remove the liquid from the slides and place the slides on a slide holder. Wash samples by allowing the slide holder to pass through glass tanks containing 1x PBS (pH 7.4) for 5 min, three times.
Briefly remove the liquid from the slides and apply 300 μl 1x PBS (pH 7.4) containing 8 μg/ml Höechst 33342 to each slide using a pipette. Gently cover the slide with a coverslip and incubate for 30 min in the dark.
Briefly remove the liquid from the slides and place them on a slide holder. Wash samples by allowing the slide holder to pass through glass tanks containing 1x PBS (pH 7.4) for 20 min, two times, then a third time overnight.
Apply 300 μl 1x PBS (pH 7.4) to the slides and gently cover each slide with a coverslip. Observe the sectioned samples using a confocal laser scanning microscope equipped with a 40x oil immersion objective. Use a 488 nm laser to excite Alexa 488 (now covalently bonded to EdU) and a 490-535 nm emission spectrum to detect Alexa 488. Use a 405 nm laser to excite Höechst 33342 and a 420-470 nm emission spectrum to detect Höechst 33342.
Store slides in a desiccated slide box at 4 °C for up to 6 months.
Data analysis
Cell pairs suitable for cell division orientation analysis are easily recognized in both Arabidopsis (Figure 3A) and J. prismatocarpus (Figure 3B) (Yin and Tsukaya, 2016 and 2019). Using these cell pairs, cell division angles can be quantified (Figure 4). The cell division angles are further categorized according to their values. In Arabidopsis, the cell division angles are categorized as follows: along the proximal-distal axis (0-30° and 150-180°), perpendicular to the proximal-distal axis (60-90° and 90-120°), and intermediate (30-60° and 120-150°). In J. prismatocarpus, the cell division angles are categorized as follows: along the thickness orientation (0-30° and 150-180°), perpendicular to the thickness orientation (60-90° and 90-120°), and intermediate (30-60° and 120-150°). From 1620 cell pairs (three samples) of Arabidopsis, leaf primordia were determined to be marked by non-stochastic cell division orientations in different localized regions (Yin and Tsukaya, 2016). From 379 cell pairs (26 samples) of J. prismatocarpus abaxialized leaf blades (Yamaguchi et al., 2010), cell divisions were found to be more frequent in the leaf thickness orientation and distributed towards the shoot apical meristem (Yin and Tsukaya, 2019).
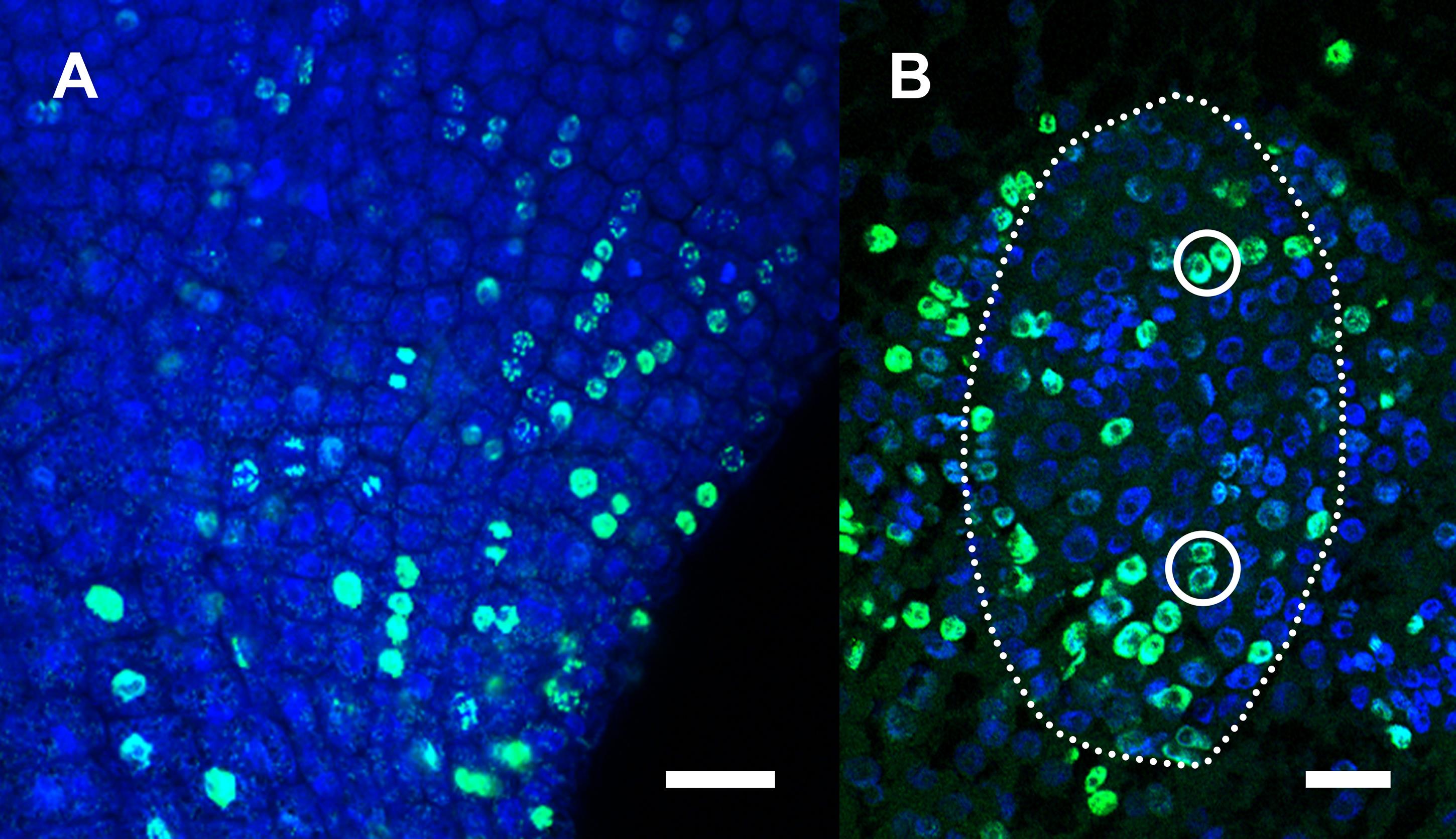
Figure 3. Representative images of detected cell pairs. A. Part of an Arabidopsis leaf primordium. B. Leaf blade of a sectioned leaf primordium (dotted outline) of Juncus prismatocarpus. Scale bars = 20 μm.
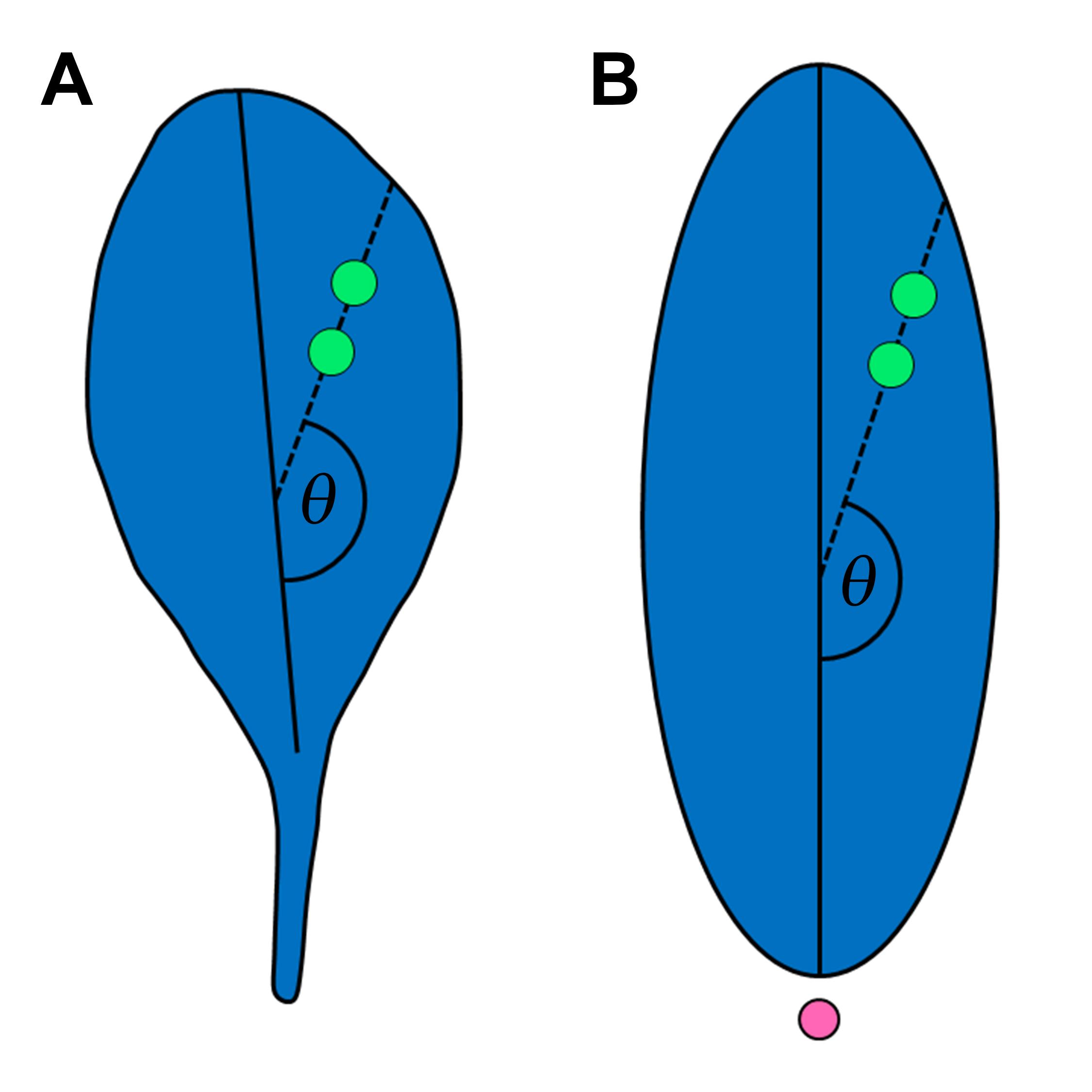
Figure 4. Cell division angle (θ). A. Schematic view of an Arabidopsis leaf. Green dots, imaginary detected cell pair. Solid line, proximal-distal axis of Arabidopsis leaf. B. Schematic view of a sectioned Juncus prismatocarpus leaf blade. Green dots, imaginary detected cell pair. Pink dot, shoot apical meristem. Solid line, thickness orientation of J. prismatocarpus leaf blade.
Notes
Preserve the cut root as a positive control, because cells in the root apical meristem are easily stained by EdU.
To ensure accurate timing when incubating many samples, add EdU to the liquid MS medium after all samples have been transferred into the tube.
If the overall EdU (Alexa 488) signal is low, the following can be performed: cut the hypocotyl at a position near the shoot apex, remove older leaves, and increase the pulse time in Steps A2 and B2.
If EdU (Alexa 488) pairs are not abundant, the following can be performed: increase the chase time in Steps A4 and B4.
When working on a new species, the pulse and chase times must be determined empirically; the times of Arabidopsis and J. prismatocarpus can be used as references. The pulse time can be determined by gradually increasing the EdU incubation time in Steps A2 and B2 until the overall EdU (Alexa 488) signal reaches a satisfactory level. The chase time can be determined by gradually increasing the chase time in 1-h increments in Steps A4 and B4 until there are sufficient EdU (Alexa 488) pairs for quantitative analysis.
The EdU detection cocktail must be made in the order of components shown in the Recipes section; it must be made fresh each time, immediately prior to detection.
Keep samples and slides in the dark (e.g., wrapped in aluminum foil) following detection by the EdU detection cocktail.
Xylene is toxic, flammable, and highly volatile. It must be handled with great care in a fume hood. Alternatives such as Histo-Clear can be used as a safe replacement for xylene.
When handling xylene, paraffin, DAPI, and Höechst 33342, gloves must be worn.
When applying a coverslip to a slide, take care not to generate bubbles on or near sections.
Recipes
MS plates
MS plant salt mixture 1 bag (~4.6 g)
Sucrose (2%, w/v) 20 g
Vitamin B5 1 ml
Agar (4%, w/v) 40 g
Dissolve the above in ~800 ml dH2O, adjust pH to ~5.8 using 1 M KOH, add dH2O to a final volume of 1,000 ml, autoclave, and allow to cool in plates
Liquid MS medium
Prepare in the same manner shown above for MS plates, but omit the use of agar
FAA
Formaldehyde 50 ml
Glacial acetic acid 25 ml
Ethanol 250 ml
dH2O 175 ml
EdU detection cocktail [made from Click-iTTM EdU Cell Proliferation Kit for Imaging, and must be prepared in the following order (indicated by numbering for clarity)]
a. Click-iTTM EdU reaction buffer (diluted from Component D with dH2O) 215 μl
b. CuSO4 aqueous solution (Component E) 10 μl
c. Alexa Fluor® 488 (mix Component B and Component C) 0.6 μl
d. Reaction buffer additive (diluted from Component F) 25 μl
Acknowledgments
We thank Professors Akihiko Nakano (The University of Tokyo, Japan) and Takashi Ueda (The University of Tokyo, Japan), for their permission to use the confocal microscope. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (Scientific Research on Priority Areas and Scientific Research on Innovative Areas, grants no. 25113001, 25113002, and 19H05672) and the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research in Japan (grant no. P19085).
Competing interests
There are no competing interests.
References
- Fukushima, K., Fujita, H., Yamaguchi, T., Kawaguchi, M., Tsukaya, H. and Hasebe, M. (2015). Oriented cell division shapes carnivorous pitcher leaves of Sarracenia purpurea. Nat Commun 6: 6450.
- Furuya, T., Hattori, K., Kimori, Y., Ishida, S., Nishihama, R., Kohchi, T. and Tsukaya, H. (2018). ANGUSTIFOLIA contributes to the regulation of three-dimensional morphogenesis in the liverwort Marchantia polymorpha. Development 145(18) .
- Horiguchi, G., Nakayama, H., Ishikawa, N., Kubo, M., Demura, T., Fukuda, H. and Tsukaya, H. (2011). ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol 52(1): 112-124.
- Ichihashi, Y., Kawade, K., Usami, T., Horiguchi, G., Takahashi, T. and Tsukaya, H. (2011). Key proliferative activity in the junction between the leaf blade and leaf petiole of Arabidopsis. Plant Physiol 157(3): 1151-1162.
- Koga, H., Doll, Y., Hashimoto, K., Toyooka, K. and Tsukaya, H. (2020). Dimorphic leaf development of the aquatic plant Callitriche palustris L. through differential cell division and expansion. Front Plant Sci 11: 269.
- Kotogany, E., Dudits, D., Horvath, G. V. and Ayaydin, F. (2010). A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6(1): 5.
- Nakayama, H., Kawade, K., Tsukaya, H. and Kimura, S. (2015). Detection of the cell proliferation zone in leaves by using EdU. Bio-protocol 5(18): e1600.
- Nakayama, H., Nakayama, N., Seiki, S., Kojima, M., Sakakibara, H., Sinha, N. and Kimura, S. (2014). Regulation of the KNOX-GA gene module induces heterophyllic alteration in North American lake cress. Plant Cell 26(12): 4733-4748.
- Nakayama, H., Yamaguchi, T. and Tsukaya, H. (2012). Acquisition and diversification of cladodes: leaf-like organs in the genus Asparagus. Plant Cell 24(3): 929-940.
- Yamaguchi, T., Yano, S. and Tsukaya, H. (2010). Genetic framework for flattened leaf blade formation in unifacial leaves of Juncus prismatocarpus. Plant Cell 22(7): 2141-2155.
- Yin, X. and Tsukaya, H. (2016). A pulse-chase strategy for EdU labelling assay is able to rapidly quantify cell division direction. New Phytol 211(4): 1462-1469.
- Yin, X. and Tsukaya, H. (2019). Morphogenesis of flattened unifacial leaves in Juncus prismatocarpus (Juncaceae). New Phytol 222(2): 1101-1111.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yin, X. and Tsukaya, H. (2021). A Pulse–chase EdU Method for Detection of Cell Division Orientation in Arabidopsis and Juncus prismatocarpus Leaf Primordia. Bio-protocol 11(1): e3882. DOI: 10.21769/BioProtoc.3882.
Category
Plant Science > Plant developmental biology > Morphogenesis
Plant Science > Plant cell biology > Cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











