- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Protocol for Simple, Rapid, and Direct Detection of SARS-CoV-2 from clinical samples, using Reverse Transcribed Loop-Mediated Isothermal Amplification (RT-LAMP)
(*contributed equally to this work) Published: Vol 10, Iss 20, Oct 20, 2020 DOI: 10.21769/BioProtoc.3789 Views: 5212
Reviewed by: Gal HaimovichShyam SolankiVaibhav B. Shah

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Colorimetric RT-LAMP Methods to Detect Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)
Gun-Soo Park [...] Jin-Soo Maeng
Nov 5, 2020 5846 Views
A Novel PCR-Based Methodology for Viral Detection Utilizing Mechanical Homogenization
Zachary P. Morehouse [...] Rodney J. Nash
Mar 5, 2022 2753 Views

Direct RNA Sequencing of Foot-and-mouth Disease Virus Genome Using a Flongle on MinION
Lizhe Xu [...] Bonto Faburay
Jun 20, 2024 2129 Views
Abstract
SARS-CoV-2 has quickly spread all around the globe causing illness and wide damages. Most countries were unprepared for such a rapid spread and crisis. This led to various strategies for effective control of the new pandemic. A key aspect in all countries was to effectively test the population for the virus. Most countries chose a lockdown strategy in which many workplaces and activities are completely closed, leading to substantial economy costs. Here, we present a protocol we recently developed that allows rapid and simple detection of SARS-CoV-2 for the large population, eliminating costs and involvement of professional teams and laboratories. This protocol is based on Reverse Transcribed Loop-Mediated Isothermal Amplification (RT-LAMP). We tested this protocol directly on patient samples, both nasal and throat clinical swabs as well as saliva. Notably, this protocol is simple, cheap and can be easily applied to other pathogens as well.
Keywords: SARS-CoV-2Background
The Covid-19 pandemic, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is affecting large populations, and has been declared a pandemic by the World Health Organization (WHO).
Mass surveillance of the population and quarantine proved to be effective strategies in dealing with this crisis. The main key for detection is the reverse transcription quantitative polymerase chain reaction (RT-qPCR) test. While this test is effective, it requires professional experience both in sampling and in performing the test. Furthermore, the reagents and lab equipment are expensive (Bruce et al., 2020). Altogether, these created a bottleneck for mass scale testing.
Fortunately, to date, alternative molecular biology methods can overcome such limitations. One of these methods is colorimetric Loop-Mediated Isothermal Amplification (LAMP) (Notomi et al., 2000). LAMP is performed at a single and constant temperature (i.e., isothermal), and allows a one-step reverse transcription. Its results can be visualized by color change with the naked eye. This method is cheap and requires little to no lab equipment (Wang et al., 2016; Yu et al., 2020; Zhang et al., 2020). It can be widely used in points of care such as the workplace and schools, which can increase the number of tested subjects tremendously. We see this method as a surveillance tool to markedly increase the total number of tests per day and identify patients for further tests in hospital settings.
Here we present a protocol for applying one step reverse transcribed LAMP (RT-LAMP) detection method on clinical samples from nasal, throat swabs and from saliva. The primers we used were previously designed by Zhang et al., 2020 on synthetic and purified RNA (Table 1). The protocol we present (Ben-Assa*, Naddaf* et al., 2020), while with these same primers, does not require RNA purification steps and can be conducted directly on clinical samples. This protocol requires no professional experience and results can be obtained within an hour. The results of this protocol were compared to the approved RNA purification and quantification RT-qPCR method at the Rambam Health Care Campus (RHCC) hospital. The downside of this method is its detection sensitivity, which is considerably lower than the standard RT-qPCR method (Ben-Assa*, Naddaf* et al., 2020). Therefore, it is applicable for a large-scale surveillance tool rather than a replacement of the current gold-standard detection method. While this protocol was validated on SARS-CoV-2, it can be adjusted to other pathogens too (bacteria or viruses).
Material and Reagents
- Personal protective equipment (PPE) appropriate for working with SARS-CoV-2
- 0.2 ml PCR tube strips (Labcon, catalogue number: LC 3927-550)
- Pipette tips
- WarmStart® Colorimetric LAMP 2x Master Mix (New England BioLabs Inc., catalog number: M1800 ) (aliquot and store at -20 °C)
- Proteinase K (Seegene, catalog number: 744300.4.UC384, store at -20 °C)
- Guanidine hydrochloride (Sigma, catalog number: G4505 )
- DNase RNase free water (Biological Industries, catalog number: 01-869-1B)
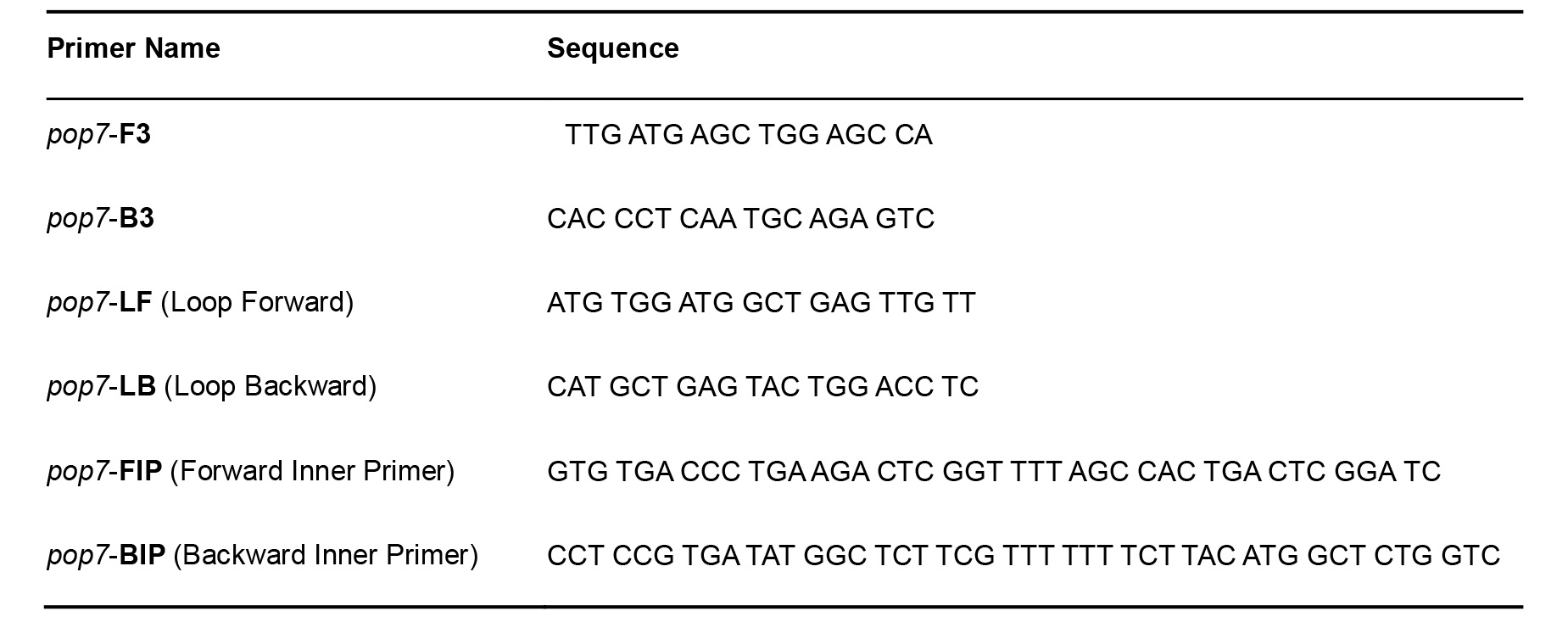
- Primers for SARS-CoV-2 (Table 1) (Zhang et al., 2020)
- Primers for pop7–positive internal processing and amplification control (Table 2) (Curtis et al., 2018)
- Primer mix (see Recipes)
- Proteinase K (see Recipes)
- Guanidine hydrochloride (see Recipes)
Table 1. SARS-CoV-2 Primers for RT-LAMP
Table 2. RNaseP pop7 Primers for RT-LAMP–Positive Control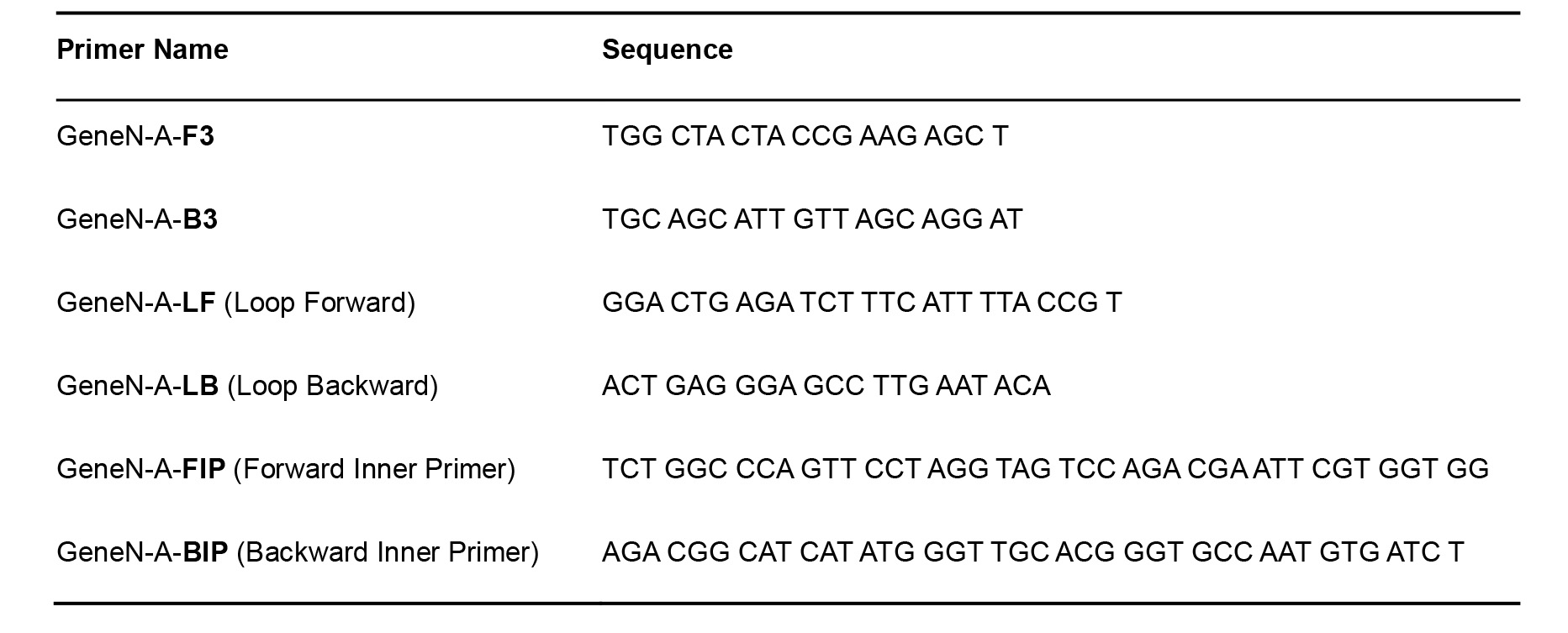
Equipment
- 1 µl-10 µl, 10 µl-50 µl, 20 µl-200 µl pipettes
- PCR Thermocycler as heat source (Biometra, catalog number: T3000 )
Procedure
- Prepare primer mix. (see Recipes)
Prepare two mixes: (1) For SARS-Cov-2 test. (2) For pop7 positive internal processing and amplification control. - Sample lysis and preparation (see Figure 1A)
- For saliva samples: Resuspend in 1 ml of DNase RNase free water (in the same container of the saliva sample). For throat/nasal swab samples: Use the standard universal transfer media (UTM) for this test.
- Add 5 µl of the sample (from Step B1) to a PCR tube containing 40 µl of DNase RNase free water.
- Include a non-template control. Add 5 µl DNase RNase free water instead of the sample.
- Add 2 µl proteinase K (final concentration 1.22 mg/ml).
- Incubate at room temperature for 15 min.
- Incubate samples at 95 °C for 5 min in a PCR machine or any other heat source.
Note: We have successfully used thermal cup and hot water. - Cool your sample down to room temperature.
- For saliva samples: Resuspend in 1 ml of DNase RNase free water (in the same container of the saliva sample). For throat/nasal swab samples: Use the standard universal transfer media (UTM) for this test.
- RT-LAMP reaction (see Figure 1A)
Prepare three mixes: (1) For SARS-CoV-2 test. (2) For pop7 gene as positive control. For each mix use the relevant primers. (3) For non-template control (see Step B3) using the SARS-CoV-2 primers.- Prepare reaction:
LAMP mix 10 µl
Primer mix 2 µl
Guanidine hydrochloride 1 µl
Sample lysate (product of Procedure B) 7 µl - Incubate at 65 °C for 30-40 min.
- Read results:
If the mix color changes from pink to yellow, the sample has tested positive (see Figure 1B).
If the mix maintains its pink color, the sample has tested negative (see Figure 1B).
Please note that the results are binary as positive or negative. Only bright yellow results are considered positive. Pink is considered negative, and any other range or gradient of color is considered negative.
Positive control results were published at Ben-Assa*, Naddaf* et al., 2020 (Figure 3a).
Figure 1. Procedure and results of SARS-CoV-2 RT-LAMP detection test. A. An illustration of the steps of the protocol: this illustration summarizes the main steps of the protocol. Illustration was created with BioRender.com. B. One negative sample (Neg S.) and one positive sample (Pos S.) before and after incubation. At t = 0 min (left panel), all samples were pink. After incubation of 30min (right panel), positive samples turned yellow while negative samples retained their pink color. Pictures are representative RT-LAMP test results of clinical diagnostic nasal and throat swabs of data published in Ben-Assa*, Naddaf* et al., 2020. Results were confirmed by conventional RT-qPCR clinical test for SARS-CoV-2 following RNA extraction and purification step.
- Prepare reaction:
Notes
- Alternative to a PCR machine, any heating source (e.g., water bath) can be used for Steps B5 and C2. We have successfully used a thermos cup and a thermometer to adjust water temperature.
- Longer incubation than 40 min may result in non-specific color change and interpreted as false positive results.
- It is very important to use proper personal protective equipment when handling the samples starting from step B and throughout the rest of the protocol.
- The color change in the protocol is an indication for a binary positive/negative result and does not represent any gradient indication of the clinical status of the patient.
Recipes
- Primer mix
- Prepare each one of the primers at 100 µM
- Primer mix:
F3 2 µl (2 µM final concentration)
B3 2 µl (2 µM final concentration)
LF 4 µl (4 µM final concentration)
LB 4 µl (4 µM final concentration)
FIP 16 µl (16 µM final concentration)
BIP 16 µl (16 µM final concentration) - DNase RNase free water 56 µl
- Aliquot and store at -20 °C
- Proteinase K at stock concentration 28.67 (mg/ml)
Soluble in DNase RNase free water
Store at -20 °C - Guanidine hydrochloride
Soluble in DNase RNase free water
Stock concentration, 800 mM
Store at room temperature
Acknowledgments
We thank the Geva-Zatorsky lab for fruitful discussions and contributions. We would like to thank the Rambam hospital for their support and for hosting us, especially to the infectious disease and virology teams. We would also like to thank Dr. Rich Roberts and Dr. Nathan Tanner for their valuable support, as well as Prof. Daniel King, Prof. Yehuda Chowers, Dr. Ronit Almog, Dr. Yuval Geffen, Dr. Dani Zvi-Bar and Prof. Oded Lewinson for their crucial help. In addition, we would like to thank Dima Abdu for preparing the figure for this paper. The illustration in this in Figure 1B was created with BioRender.com.
This work was supported by the Technion Integrated Cancer Center, the Technion – Israel Institute of Technology, “Keren Hanasi”, Alon Fellowship for Outstanding Young Researchers, Horev Fellow (Taub Foundation). NGZ is an Azrieli Global Scholar at the Canadian Institute for advanced research (CIFAR).
Competing interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. While performing this research, no company had invested in the research, and no commercialization was intended.
Ethics
This study was granted exemption from IRB approval of the Rambam Health Care Campus (# 0244-20-RMB) for use of de-identified COVID-19 tests performed for the purpose of the standard testing, and for 4 volunteers.
References
- Ben-Assa, N., Naddaf, R., Gefen, T., Capucha, T., Hajjo, H., Mandelbaum, N., Elbaum, L., Rogov, P., King, D. A., Kaplan, S., Rotem, A., Chowers, M., Szwarcwort-Cohen, M., Paul, M. and Geva-Zatorsky, N. (2020). Direct on-the-spot detection of SARS-CoV-2 in patients. Exp Biol Med (Maywood) 245(14): 1187-1193.
- Bruce, E. A., Huang, M. L., Perchetti, G. A., Tighe, S., Laaguiby, P., Hoffman, J. J., Gerrard, D. L., Nalla, A. K., Wei, Y., Greninger, A. L., Diehl, S. A., Shirley, D. J., Leonard, D. G. B., Huston, C. D., Kirkpatrick, B. D., Dragon, J. A., Crothers, J. W., Jerome, K. R. and Botten, J. W. (2020). DIRECT RT-qPCR DETECTION OF SARS-CoV-2 RNA FROM PATIENT NASOPHARYNGEAL SWABS WITHOUT AN RNA EXTRACTION STEP. bioRxiv. doi: 10.1101/2020.03.20.001008.
- Curtis, K. A., Morrison, D., Rudolph, D. L., Shankar, A., Bloomfield, L. S. P., Switzer, W. M. and Owen, S. M. (2018). A multiplexed RT-LAMP assay for detection of group M HIV-1 in plasma or whole blood. J Virol Methods 255: 91-97.
- Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., and Hase, T. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12): e63-e63.
- Wang, X., Yin, F., Bi, Y., Cheng, G., Li, J., Hou, L., Li, Y., Yang, B., Liu, W. and Yang, L. (2016). Rapid and sensitive detection of Zika virus by reverse transcription loop-mediated isothermal amplification. J Virol Methods 238: 86-93.
- Yu, L., Wu, S., Hao, X., Dong, X., Mao, L., Pelechano, V., Chen, W. H. and Yin, X. (2020). Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform. Clin Chem 66(7): 975-977.
- Zhang, Y., Odiwuor, N., Xiong, J., Sun, L., Nyaruaba, R. O., Wei, H., and Tanner, N. A. (2020). Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP. MedRxiv doi: https://doi.org/10.1101/2020.02.26.20028373.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Naddaf, R., Ben-Assa, N., Gefen, T., Capucha, T., Hajjo, H., Mandelbaum, N., Elbaum, L., Kaplan, S., Rotem, A., Chowers, M., Szwarcwort-Cohen, M., Paul, M. and Geva-Zatorsky, N. (2020). A Protocol for Simple, Rapid, and Direct Detection of SARS-CoV-2 from clinical samples, using Reverse Transcribed Loop-Mediated Isothermal Amplification (RT-LAMP). Bio-protocol 10(20): e3789. DOI: 10.21769/BioProtoc.3789.
Category
Microbiology > Pathogen detection > RT-LAMP
Molecular Biology > RNA > RNA detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











