- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Single Cell Migration Assay Using Human Breast Cancer MDA-MB-231 Cell Line
Published: Vol 10, Iss 8, Apr 20, 2020 DOI: 10.21769/BioProtoc.3586 Views: 7359
Reviewed by: Gal HaimovichChaitali BasolePhilipp Wörsdörfer

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Imaging of Human Cancer Cells in 3D Collagen Matrices
Karin Pfisterer [...] Maddy Parsons
Jan 20, 2021 6199 Views

Studying Chemotactic Migration in Dunn Chamber: An Example Applied to Adherent Cancer Cells
Khedidja Benseddik and Kossay Zaoui
Feb 5, 2022 2892 Views
Image-based Quantification of Macropinocytosis Using Dextran Uptake into Cultured Cells
Anh H. Le and Laura M. Machesky
Apr 5, 2022 4099 Views
Abstract
Cell migration is a fundamental cellular process that plays a crucial role in many physioglogical and pathological processes such as wound healing or cancer metastasis. Many assays have been developed to examine cell migration, such as the wound healing or scratch assay, Boyden Chamber or transwell assay, and the method we will describe here, single cell migration assay. In this assay, cells are plated sparsely on a collagen coated plate and live cell imaging is performed over a period of 2 h at 1 frame per minute. After imaging is completed, cells are tracked manually using ImageJ by tracking movement of the centroid of the cell. These data points are then exported and overall distance travelled from frame to frame is determined and divided by total time imaged to determine speed of the cell. This method provides a quick way to examine effect of cellular manipulation on cell migration before proceeding to perform more complex assays.
Keywords: Single cell migrationBackground
Cell migration plays an important role in both physiological and pathological processes ranging from embryonic development to angiogenesis and tumor metastasis (Le Clainche and Carlier, 2008). Cell motility is a highly orchestrated event that can be summarized as a cycle of four basic steps: i) membrane protrusion driven by actin polymerization, ii) stabilization of protrusion through integrin-mediated cell-matrix adhesion, iii) cell-body translocation driven by actomyosin contractile force, and finally, iv) rear release as a result of the mechanical action of contractile force and/or proteolysis of cell-matrix adhesion components (Sheetz et al., 1999; Ridley et al., 2003; Panetti et al., 2004; Stradal and Scita, 2006; Tomasevic et al., 2007; Le Clainche and Carlier, 2008). These processes involve dynamic remodeling of actin cytoskeleton which is dependent on de novo synthesis as well as regulation of important structural and regulatory components of actin cytoskeletal system.
Several methods have been developed to examine cell migration and can be separated into 2D and 3D assays. 2D migration assay have their advantages as they are typically easier and quicker to perform, however, lack the physiological representation that 3D assays may provide. We will provide a brief description of some of the other methods for cell migration. The scratch assay or wound healing assay is a commonly used technique to evaluate directed cell migration (Cory, 2011). In this assay, cells are plated to form a confluent monolayer and a stratch is introduced in the monolayer. The cell-free zone is then monitored as adjacent cells migrate to close the gap. While simple to perform, cell proliferation may bias the readout of this assay as that can influence scratch closure. In addition, cells in this assay are forced to move in one direction. The Boyden Chamber assay or transwell assay is another technique used to evaluate cell migration (Falasca et al., 2011). In this assay, cells are placed on one side of a porous membrane and allowed to migrate through the pores to the other side. An advantage of this assay is it allows for chemotaxis and works for both adherent and non-adherent cells. As with the scratch assay, migration in this assay is defined. Some disadvantages of this assay include difficulty to visualize cells and morphology due to transitive state of cells while migrating through pores.
In this protocol, we will describe the use of single cell migration assay to examine MDA-MB-231 cell migration. The key difference in this protocol is that we examine random cell migration rather than directed migration. This allows us to examine cell migration in a manner that is less influenced by cell-cell contact and its innate ability for directional persistence, i.e., ability to migrate randomly in a single direction. While obviously not a true physiological representation of in vivo cell migration, this assay allows us to quickly analyze effect of cellular manipulation on its ability to migrate. While this assay uses MDA-MB-231, this protocol can be performed with any migratory cell line. We have performed this assay using multiple cell lines with minor modifications (media used and amount of cells plated).
Materials and Reagents
- 24-well cell culture plate (Corning, Costar, catalog number: 9761146 )
- 100 mm cell culture plate (Corning, catalog number: 08-772-22 )
- 15 ml conical centrifuge tubes (Falcon, catalog number: 14-959-49D )
- 100% CO2 Tank
- DMEM (Lonza, BioWhittaker, catalog number: BW12-604F )
- Heat-inactivated FBS (Corning, catalog number: 35011CV )
- 100x Antibiotic-Antimycotic (Gibco, catalog number: 15240-062 )
Note: This is optional. We use this as extra assurance to prevent bacterial and fungal contamination in our cell culture. For some cell lines, this can hinder cell growth. - 100x Sodium pyruvate (Gibco, catalog number: 11360-070 )
- Collagen I, 3 mg/ml (Gibco, catalog number: A1048301 )
- Trypsin (Lonza, Trypsin-Versene, catalog number: BW17161E )
- PBS (Lonza, BioWhittaker, catalog number: BW17516F )
- MDA-MB-231 (ATCC, catalog number: CRM-HTB-26 )
- DMEM S+ media (see Recipes)
- Collagen (10 μg/ml) (see Recipes)
Equipment
- Hemocytometer
- Live-cell imaging environmental chamber (Tokai Hit live-cell environmental chamber)
- Centrifuge (unrefrigerated) for 15 ml tubes (Sorvall, Legend RT, SO-LEGRT)
- Cell incubator set to 37 °C and 5% CO2
- Waterbath or drybath set to 37 °C (Boekel Scientific, Small Water Bath, 290100)
- Microscope with automatic stage control (Olympus IX71 Inverted Phase with Prior Scientific motorized stage) (Figure 1)
- Camera for microscope (Hamamatsu)
- 10x objective (Olympus)

Figure 1. Example of a complete setup for cell tracking using Olympus IX71 inverted microscope, Prior mechanized stage, and Tokai Hit environmental chamber
Software
- Software compatible with microscope for time-lapse image aquisition (Olympus, cellSens)
- Fiji ImageJ (https://fiji.sc/)
- Microsoft Excel
Procedure
- Cell culture
- Warm up DMEM S+ media to 37 °C.
- Thaw MDA-MB-231 cells to 37 °C.
- Transfer 1 ml of thawed MDA-MB-231 cells to 3 ml of DMEM S+ media and centrifuge (with appropriate balance) for 3 min at 300 x g.
- Aspirate supernatant and resuspend cells in MDA-MB-231 cells in 10 ml of DMEM S+ and plate on 10 cm dish.
- Incubate cells at 37 °C and 5% CO2 until 70% confluency (2-3 days).
- Single cell migration assay
- Warm up DMEM S+ media, trypsin, and PBS to 37 °C.
- Coat 24-well plate with 10 μg/ml Collagen-I solution (100 μl) for 1 h at room temperature and wash with PBS (leave PBS in well to prevent drying).
- Aspirate media from MDA-MB-231 cell culture dish.
- Wash cells with PBS and aspirate PBS.
- Add 1 ml of trypsin and incubate in 37 °C and 5% CO2 for 5 min.
- Pick up trypsinized cells with 3 ml of DMEM S+ and centrifuge for 3 min with appropriate balance at 300 x g.
- Aspirate supernatant and resuspend cells in 4 ml of DMEM S+.
- Take 10 μl of cell suspension and pipette into hemocytometer.
- Count cells.
- Aspirate PBS off collagen coated 24-well plate.
- Resuspend cell suspension again by inverting tube a few times or pipetting.
- Plate 20,000 cells into 24-well and allow to attach overnight at 37 °C and 5% CO2.
- Allow live-cell imaging environmental chamber warm up on microscope stage with empty 24-well plate for at least 1 h to equilibrate microscope temperature.
- Replace empty plate with cell plated 24-well and allow to equilibrate for 1 h.
- Select multiple fields with 5-10 single cells (not touching other cells) and set time-lapse imaging for 2 h at 1 frame per minute using phase-contrast.
- Initiate time-lapse imaging and save files.
Data analysis
- If fields were saved as individual images (i.e., 1 image per field per time point), import image sequence on Fiji to build a montage of all images from one field. If fields were saved as an individual file (i.e., 1 file per field), open directly with Fiji.
- Play through the movie and identify cells that do not touch other cells or migrate off the field of view (try to identify 5-10 cells or more per field is possible). See Video 1 for example field of migrating cells.Video 1. Example video of cell migration from Figure 2
- Once candidate cells have been determined, open Manual Tracking plugin on Fiji (Figure 2A, a window should open).
- Click “add track” (Figure 2B).
- Begin clicking on the centroid of cell (center of cell) and continue tracking until all 120 frames have been tracked (Figure 2C). Note that data will be added to a new window (Figure 2D).
- Click “end track” and close manual tracking window (Figure 2E).
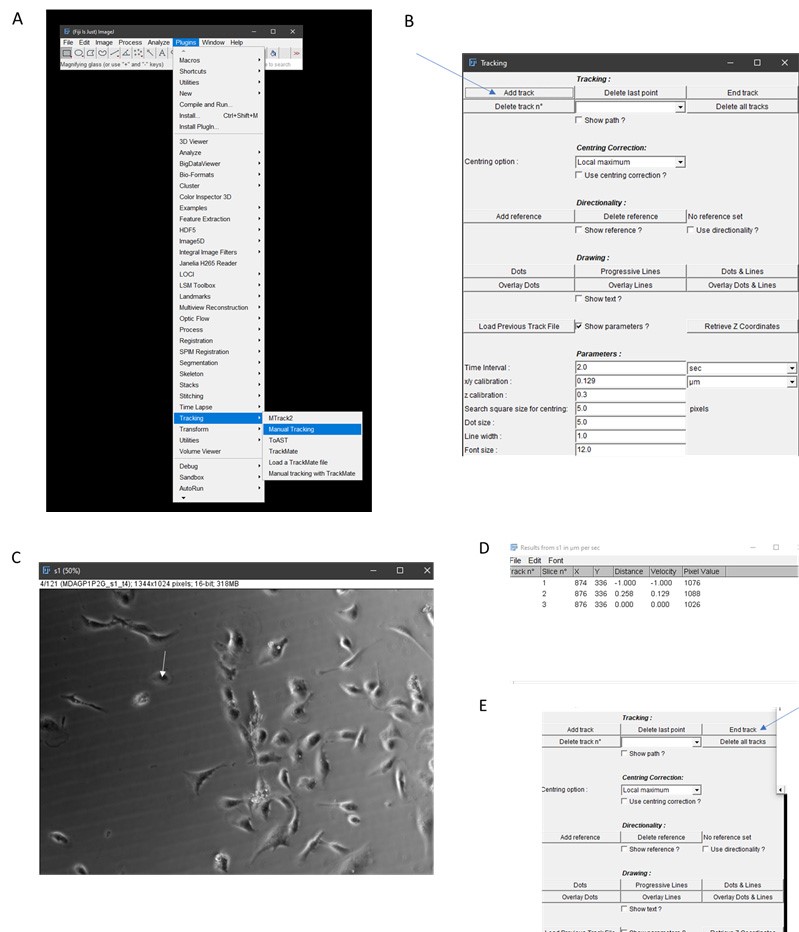
Figure 2. Workflow for manual tracking using Fiji. A. Select Manual Tracking under Plugins. B. Click Add Track. C. Begin tracking a cell (continuously click on centroid of cell, for example, as illustrated by white arrow. Note that cells that are clumped together like those in the middle of this panel should not be used for tracking. D. Tracking information will be generated by ImageJ. E. Click End Track when tracking is done. - Save results window which contains X, Y coordinates of the cell centroid per frame.
- Repeat Steps 3-7 until all cells have been quantified and proceed to next field.
- Once all cells and fields have been quantified, open Excel and paste X,Y coordinates (Figure 3A).
- Determine distance travelled from point to point by using the distance formula below where X1 and Y1 are the current track location and X2 and Y2 are the next track location. Find d for each tracked point (Figure 3B, there will be 121 tracked points, since there is no previous point to the first tracked spot, you will end up with 120 calculations for d).

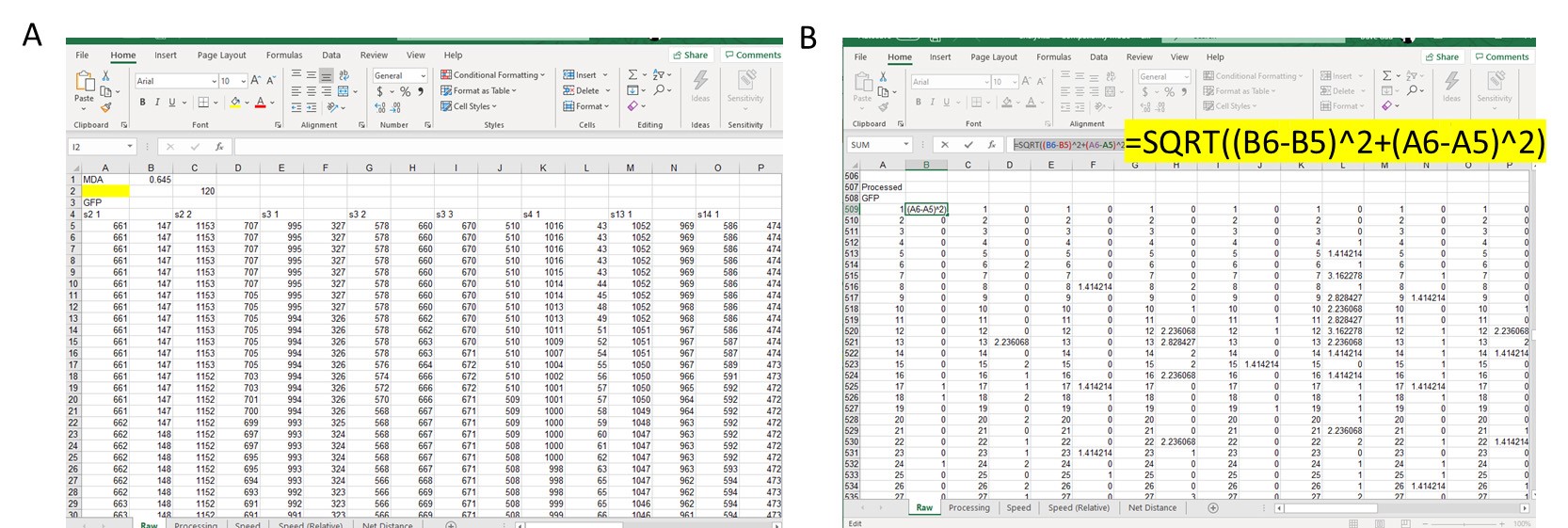
Figure 3. Example of data analysis using Excel. A. Copy over X and Y coordinates from cell tracking into Excel and line up side-by-side for each cell tracked. B. In separate section, use the distance formula to calculate distance travelled by cell from Point 1 to Point 2, Point 2 to Point 3, Point 3 to Point 4, and so on. Example of Excel function used is highlighted. - Add up all distance travelled per cell and divide by 120 (this is the duration of the movie in min and also the number of frames per field).
- Repeat Steps 10 and 11 for all cells.
- Summarize cell speed as desired and perform appropriate statistical analysis (t-test for 2 groups, ANOVA with post hoc for > 2 groups).
- Plot summarized information using preferred graph (i.e., bar graph, box-whisker, scatter plot). An example of a box-whisker plot is shown in Figure 4.
Figure 4. Example of speed results plotted as a box and whiskers chart using Excel. G1, G2, G3 are generic group names. The blue star represents the mean and the green bar represents the median of the data.
Recipes
- DMEM S+ media
500 ml DMEM
50 ml FBS
5 ml 100x Antibiotic-Antimycotic
5 ml 100x Sodium Pyruvate - Collagen (10 μg/ml)
10 ml DMEM
166.6 μl Collagen I 3 mg/ml
Acknowledgments
This work was supported by a grant from the National Institute of Health (2R01CA108607) to PR. David Gau was supported by a National Science Foundation pre-doctoral fellowship (2012139050) and an NIH Cardiovascular Bioengineering pre-doctoral training grant (2T32HL076124 to SG).
Competing interests
The authors have no competing interests to report.
References
- Cory, G. (2011). Scratch-wound assay. Methods Mol Biol 769: 25-30.
- Falasca, M., Raimondi, C. and Maffucci, T. (2011). Boyden chamber. Methods Mol Biol 769: 87-95.
- Le Clainche, C. and Carlier, M. F. (2008). Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev 88(2): 489-513.
- Panetti, T. S., Hannah, D. F., Avraamides, C., Gaughan, J. P., Marcinkiewicz, C., Huttenlocher, A. and Mosher, D. F. (2004). Extracellular matrix molecules regulate endothelial cell migration stimulated by lysophosphatidic acid. J Thromb Haemost 2(9): 1645-1656.
- Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Ginsberg, M. H., Borisy, G., Parsons, J. T. and Horwitz, A. R. (2003). Cell migration: integrating signals from front to back. Science 302(5651): 1704-1709.
- Sheetz, M. P., Felsenfeld, D., Galbraith, C. G. and Choquet, D. (1999). Cell migration as a five-step cycle. Biochem Soc Symp 65: 233-243.
- Stradal, T. E. and Scita, G. (2006). Protein complexes regulating Arp2/3-mediated actin assembly. Curr Opin Cell Biol 18(1): 4-10.
- Tomasevic, N., Jia, Z., Russell, A., Fujii, T., Hartman, J. J., Clancy, S., Wang, M., Beraud, C., Wood, K. W. and Sakowicz, R. (2007). Differential regulation of WASP and N-WASP by Cdc42, Rac1, Nck, and PI(4,5)P2. Biochemistry 46(11): 3494-3502.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Gau, D. M. and Roy, P. (2020). Single Cell Migration Assay Using Human Breast Cancer MDA-MB-231 Cell Line. Bio-protocol 10(8): e3586. DOI: 10.21769/BioProtoc.3586.
- Gau, D., Veon, W., Shroff, S. G. and Roy, P. (2019). The VASP-profilin1 (Pfn1) interaction is critical for efficient cell migration and is regulated by cell-substrate adhesion in a PKA-dependent manner. J Biol Chem 294(17): 6972-6985.
Category
Cancer Biology > Invasion & metastasis > Cell biology assays > Cell migration
Cell Biology > Cell movement > Cell migration
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link