- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fibroblast Gap-closure Assay-Microscopy-based in vitro Assay Measuring the Migration of Murine Fibroblasts
Published: Vol 9, Iss 16, Aug 20, 2019 DOI: 10.21769/BioProtoc.3333 Views: 5836
Reviewed by: Meenal SinhaPaula clarisa EllenbergAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Lipid-Mediated Sequential Recruitment of Proteins Via Dual SLIPT and Dual SLIPTNVOC in Live Cells
Kristina V. Bayer and Richard Wombacher
Nov 5, 2025 1580 Views
Utilizing EdU to Track Leukocyte Recruitment to the Brain
Zoie K. Lipfert [...] David P. Sullivan
Dec 5, 2025 1536 Views
Abstract
Pulmonary fibrosis is characterized by pathological scaring of the lung. Similar to other fibrotic diseases, scar formation is driven by excessive extracellular matrix deposition by activated, proliferative, and migratory fibroblasts.
Currently, the two most widely used chemotaxis and cell migration assays are the scratch assay and the transmembrane invasion assay. Here we present a gap closure assay that employs commercially available cell lines, equipment and reagents and is time efficient as well as straightforward. The protocol uses an Oris pro cell migration assay 96-well plate with a dissolvable plug in the center of each well to create a cell free area at the time of seeding. Cell repopulation of the empty zone is captured via light microscopy at different time points and quantified with free image analysis software. The clear advantages of this assay in comparison to similar protocols are the use of uncomplicated cell culture methods and the ability to image the experiment throughout.
Background
Few treatments for fibrosing diseases exist because of an incompletely understood and complex etiology (Rockey et al., 2015). Current efforts to develop therapies for organ fibrosis have focused on pathologic fibroblasts, also known as myofibroblasts (Blackwell et al., 2014). In addition to secretion of matrix proteins such as collagen, which comprise scar, a hallmark of pathologic fibroblasts is their increased proliferative and migratory capacity (Kendall and Feghali-Bostwick, 2014). A number of studies have shown that innate immune cells interact with and mediate fibroblast activation (Desai et al., 2018). Thus, we investigated the effects of lung macrophages on fibroblasts in a recent study, where we found a novel population of macrophages that localizes to fibrotic scar in murine lung (Aran et al., 2019). These macrophages highly expressed PDGF-AA, a secreted factor known for promoting fibroblast migration and proliferation. Thus, we treated mouse 3T3 embryonic fibroblasts with conditioned media from cultured mouse lung macrophages, with and without PDGF-AA blocking antibody, and measured fibroblast gap closure.
Here we describe the protocol for the assay, which should be useful for other investigators studying paracrine signaling between adjacent cellular lineages. Since the method presented employs an established, adherent cell line, it may be easily adapted to other cell types and treatments in fields where cell migration is an important pathological characteristic, such as cancer or wound healing. Importantly, fibroblasts and the extracellular matrix have been recently recognized as fundamental players in the tumor microenvironment and as such are of the outmost interest in oncology research (Bu et al., 2019).
Compared with other methods for investigating chemotaxis (Justus et al., 2014), such as the scratch assay, our approach does not introduce mechanical stress to the cells, which can potentially activate fibroblasts and obscure results. It also does not require optimization of cell culture as required for the transwell/chamber invasion assay, and data from the same well may be collected at multiple timepoints while cells are visualized in real time. Our assay can be easily scaled up to a high throughput format for drug screening purposes. One limitation of the assay is that it does not address directional cell migration. Also, our protocol measures both migration and proliferation of fibroblast cells; a proliferation assay should be conducted if further distinction is needed.
Materials and Reagents
- Oris pro cell migration assay plate, 96-well (Platypus Technologies, catalog number: PROCMA1), stored at room temperature
- 3T3 cells (ATCC, catalog number: CRL-1658), stored in liquid nitrogen
- DMEM (Corning, catalog number: 10-101-CV), stored at 4 °C
- Fetal Bovine Serum (HyClone, catalog number: SH3039603LR), stored at 4 °C
- Antibiotic-antimycotic solution (Corning, catalog number: 30-004-CI), stored at 4 °C
- Trypsin 0.25% (Corning, catalog number: 25053CL), stored at 4 °C
- PDGF-AA antibody (Millipore, catalog number: 07-1436), stored at -20 °C
- Cell culture media (see Recipes)
- Fully supplemented media (see Recipes)
- Serum free media (see Recipes)
Equipment
- Microscope
Zeiss Axio Observer D1 (Carl Zeiss) equipped with a Yocogawa spinning wheel coupled to a photometrics EMCCD camera (Evolve 512 delta)
Software
- Zeiss Zen Blue (Carl Zeiss)
- Fiji/ImageJ (https://fiji.sc/)
- Prism (https://www.graphpad.com/scientific-software/prism/)
Procedure
- Establish healthy culture of 3T3 cells in a fully supplemented media (10% serum) according to instructions from ATCC.
- On the day of the experiment, trypsinize the cells, count and re-suspend in a pre-warmed serum-free media (100 μl per well). Add approximately 5 x 104 cells per well (100% confluent) to a migration assay plate; this step may need optimization depending on cell size and doubling time. Plan to have 3 wells per condition. Let the cells to adhere for 3 h. Note that the plaque in the center of the well dissolves within 30 min; therefore, it is suitable only for rapidly adherent cells. For example, we found that freshly sorted primary mouse lung fibroblasts needed a longer time to adhere, and we failed to obtain consistent results from primary cells.
- Using the light microscope (phase contrast for unlabeled cells), verify the cell attachment and the presence of a circular cell-free zone in each well. Exclude the wells that have an excessive amount of cells in a cell-free zone or have uneven distribution of cells in cell growth area (approximately 5% of wells). Take the 0 h timepoint picture of each well, remembering to number or name the wells and images. We recommend using a microscope with a built-in incubator chamber to avoid additional stress to the cells.
- Add 100 μl of pre-warmed media with antibodies/drug or conditioned media. Make sure that all of the wells have the same serum conditions. For example, our conditioned media from mouse macrophages contained 10% serum; we therefore used 10% serum media for blocking antibody and all control wells.
- Acquire images at 24 h and, if needed, at later timepoints for each well. We found that at 48 h the gaps in majority of wells were completely closed and the most significant difference of the gap closure occurred during the first 24 h. It is possible that shorter timepoints might be necessary for other cell types, or when using conditions with potent mitogens.
Data analysis
We used Zeiss microscope software Zen Blue to stitch the phase-contrast images of the wells acquired at 20x magnification. Fiji (ImageJ) was used for all further image processing and data analysis. As seen in Figure 1, inverting the colors of the pictures greatly improved clarity of the cell-free zone border. We manually selected the cell-free zones using the circular selection tool for 0 h timepoint images and the circular or manually drawn (if the area was not circular) selection tool for 24 h timepoint images. We took individual pictures of wells at time 0 h, because we noticed slight size variability of the cell free zones between the wells. Next, the area of selection was measured by implementing the commands in Fiji: analyze > measure > compute area. We calculated the migration area as a difference between area at time 0 h and 24 h for each well, averaged the area for triplicate wells, and then normalized the data (i.e., we divided all data points from all groups by a mean of the ‘baseline’ group). We used two-sided Wilcoxon rank-sum test as the statistical test.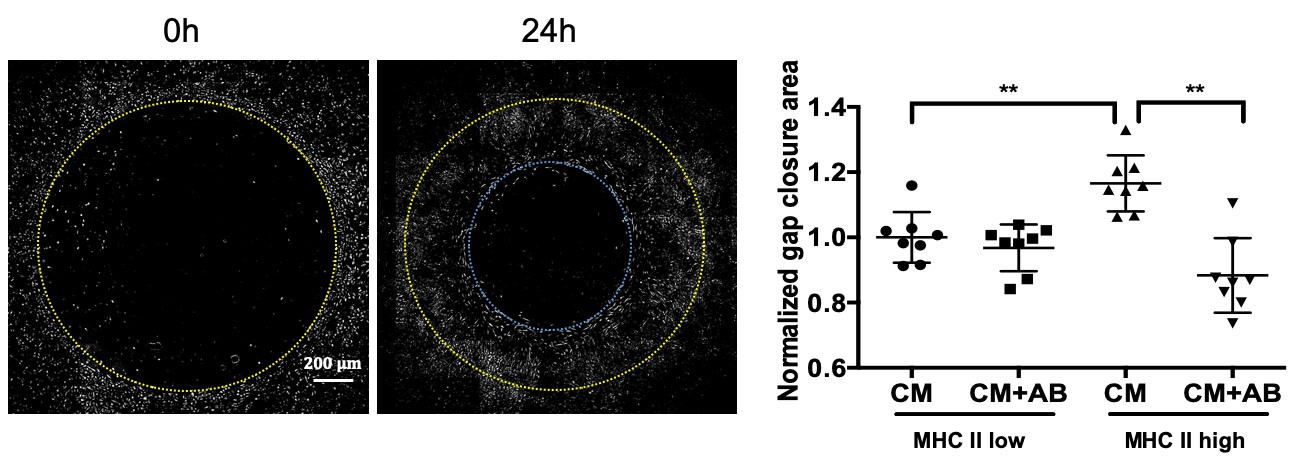
Figure 1. Representative images and quantification of fibroblast gap closure assay. Pictures of 3T3 fibroblasts were taken at time 0 h and 24 h. Colors in the image were inverted for clarity, the yellow circle indicates the border of the cell free zone at time 0 h and blue circle at time 24 h. Representative data reproduced from Aran et al., 2019; 3T3 fibroblasts were incubated with conditioned media (CM) from lung macrophages sorted by MHCII expression, with and without PDGF-AA blocking antibody (AB). Wilcoxon test two-sided P values are presented. **P < 0.01.
Recipes
- Cell culture media
DMEM
Fetal Bovine Serum
Antibiotic-antimycotic solution
Trypsin
Prepare cell culture media in sterile conditions - Serum free media
DMEM
1% antibiotic-antimycotic - Fully supplemented media
DMEM
10% serum
1% antibiotic-antimycotic
Acknowledgments
This work was supported by a UCSF Marcus Award and a National Institutes of Health grant (HL131560) to Mallar Bhattacharya.
Our assay uses commercially available Oris Pro Migration Assay (https://www.platypustech.com/cell-migration), which has been optimized and modified for this protocol.
Competing interests
The authors declare no competing financial interest.
References
- Aran, D., Looney, A. P., Liu, L., Wu, E., Fong, V., Hsu, A., Chak, S., Naikawadi, R. P., Wolters, P. J., Abate, A. R., Butte, A. J. and Bhattacharya, M. (2019). Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20(2): 163-172.
- Blackwell, T. S., Tager, A. M., Borok, Z., Moore, B. B., Schwartz, D. A., Anstrom, K. J., Bar-Joseph, Z., Bitterman, P., Blackburn, M. R., Bradford, W., Brown, K. K., Chapman, H. A., Collard, H. R., Cosgrove, G. P., Deterding, R., Doyle, R., Flaherty, K. R., Garcia, C. K., Hagood, J. S., Henke, C. A., Herzog, E., Hogaboam, C. M., Horowitz, J. C., King, T. E., Jr., Loyd, J. E., Lawson, W. E., Marsh, C. B., Noble, P. W., Noth, I., Sheppard, D., Olsson, J., Ortiz, L. A., O'Riordan, T. G., Oury, T. D., Raghu, G., Roman, J., Sime, P. J., Sisson, T. H., Tschumperlin, D., Violette, S. M., Weaver, T. E., Wells, R. G., White, E. S., Kaminski, N., Martinez, F. J., Wynn, T. A., Thannickal, V. J. and Eu, J. P. (2014). Future directions in idiopathic pulmonary fibrosis research. An NHLBI workshop report. Am J Respir Crit Care Med 189(2): 214-222.
- Bu, L., Baba, H., Yoshida, N., Miyake, K., Yasuda, T., Uchihara, T., Tan, P. and Ishimoto, T. (2019). Biological heterogeneity and versatility of cancer-associated fibroblasts in the tumor microenvironment. Oncogene 38(25): 4887-4901.
- Desai, O., Winkler, J., Minasyan, M. and Herzog, E. L. (2018). The role of immune and inflammatory cells in idiopathic pulmonary fibrosis. Front Med (Lausanne) 5: 43.
- Justus, C. R., Leffler, N., Ruiz-Echevarria, M. and Yang, L. V. (2014). In vitro cell migration and invasion assays. J Vis Exp (88). doi: 10.3791/51046.
- Kendall, R. T. and Feghali-Bostwick, C. A. (2014). Fibroblasts in fibrosis: novel roles and mediators. Front Pharmacol 5: 123.
- Rockey, D. C., Bell, P. D. and Hill, J. A. (2015). Fibrosis--a common pathway to organ injury and failure. N Engl J Med 373(1): 96.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Looney, A. P. and Bhattacharya, M. (2019). Fibroblast Gap-closure Assay-Microscopy-based in vitro Assay Measuring the Migration of Murine Fibroblasts. Bio-protocol 9(16): e3333. DOI: 10.21769/BioProtoc.3333.
Category
Cell Biology > Cell movement > Cell migration
Immunology > Inflammatory disorder > Lung injury
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link