- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Radioactive in vitro ERK3 Kinase Assay
Published: Vol 9, Iss 16, Aug 20, 2019 DOI: 10.21769/BioProtoc.3332 Views: 6733
Reviewed by: Chiara AmbrogioMauro Sbroggio'Enrico Patrucco

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Tissue-Specific Profiling of O-GlcNAcylated Proteins in Drosophila Using TurboID-CpOGAM
Qin Lei [...] Kai Yuan
Mar 5, 2025 2220 Views

Detection and Analysis of S-Acylated Proteins via Acyl Resin–Assisted Capture (Acyl-RAC)
Dina A. Abdulrahman and Michael Veit
Apr 5, 2025 1884 Views

Dual Phospho-CyTOF Workflows for Comparative JAK/STAT Signaling Analysis in Human Cryopreserved PBMCs and Whole Blood
Ilyssa E. Ramos [...] James M. Cherry
Nov 20, 2025 2379 Views
Abstract
Mitogen-activated protein kinases (MAPKs) are serine/threonine kinases that have an important role in signal transduction. Extracellular signal-regulated kinase 3 (ERK3), also known as MAPK6, is an atypical MAPK. Here, we describe in detail an in vitro assay for the kinase activity of ERK3 using myelin basic protein (MBP) or steroid receptor coactivator-3 (SRC-3) as substrates. The assay is carried out in the presence of [γ-32P]-ATP which results in radiolabeling of phosphorylated substrates. Separation of the reaction components by gel electrophoresis followed by autoradiography enables detection of the radiolabeled products, and hence determination of the kinase activity of ERK3. This assay can be used for several applications including identification of substrates, determination of the effect of molecules or mutations on kinase activity, and testing specific kinase inhibitors. Furthermore, the protocol outlined here can be adapted to measure the activity of other kinases by using their specific substrates.
Keywords: ERK3Background
Extracellular signal-regulated kinase 3 (ERK3) is an atypical mitogen-activated protein kinase (MAPK) (Coulombe and Meloche, 2007). Here we describe an in vitro kinase assay in which ERK3 transfers radiolabeled gamma-phosphate from [γ-32P]-ATP to a purified protein substrate. Excess radiolabeled ATP is then separated from the radiolabeled substrate by gel electrophoresis. The amount of phosphorylated substrate can be quantified by autoradiography, phosphorimaging, or liquid scintillation counting techniques. Radioactive kinase assay provides a direct measurement of kinase activity. It is sensitive, quick, inexpensive, and considered as the 'gold standard' for quantification of protein kinase activity. The major limitations of radioactive kinase assays are the hazards of handling radiolabeled isotopes and unsuitability of this assay format for large scale high-throughput screening.
This protocol describes a direct kinase assay for ERK3 using myelin basic protein (MBP) or a fragment of steroid receptor coactivator-3 (SRC-3) as substrates. MBP is a non-specific substrate for several kinases including members of MAPK family (Haubrich and Swinney, 2016). SRC-3 was shown to interact with ERK3 and is phosphorylated by ERK3 on its Ser857 residue within the CBP-interacting domain (CID) (Long et al., 2012). Hence, SRC3-CID fragment, which comprises amino acids 841-1080, is used as a substrate in this assay.
Another substrate for ERK3 that has been well-characterized and validated to be physiologically relevant is MAPK-activated protein kinase 5 (MK5) (Schumacher et al., 2004, Seternes et al., 2004). ERK3 phosphorylates MK5 at Thr182, leading to MK5 activation. Since MK5 itself is also a kinase, the activity of ERK3 towards MK5 has been determined by a coupled kinase assay in which the phosphorylation of peptide or protein substrate for MK5 is measured in the presence of both ERK3 and MK5 (Schumacher et al., 2004, Seternes et al., 2004).
The in vitro kinase assay described here has been previously used to identify novel ERK3 substrates (Bian et al., 2016), to determine the effect of mutations on the kinase activity of ERK3 (Alsaran et al., 2017, Elkhadragy et al., 2018), and to compare autophosphorylation of wild type or mutant ERK3 (Elkhadragy et al., 2018). In these studies, ERK3 protein was expressed and purified from bacteria, Sf9 insect cells, or mammalian cells. HA-tagged ERK3 expressed and immunoprecipitated from mammalian 293T cells showed greater in vitro kinase activity as compared to recombinant His-tagged ERK3 purified from E. coli (Elkhadragy et al., 2018), possibly because of the greater extent of post-translational modifications or the presence of interacting partners in mammalian cells. Selection of the method for purifying ERK3 protein to be used in a kinase assay has to be based on the purpose and specific considerations of the experiment to be conducted.
Materials and Reagents
- Pipette tips
- Eppendorf tubes
- X-ray film
- Filter paper (Bio-Rad Laboratories, catalog number: 1703965)
- ERK3 protein: Wild type or mutant ERK3 proteins can be purified from Sf9 insect cells, mammalian cells, or E. coli as described previously (Bian et al., 2016, Elkhadragy et al., 2018)
- Recombinant protein substrates: GST-SRC3-CID can be purified as described previously (Elkhadragy et al., 2018), and Myelin Basic Protein (MPB) is commercially available (Millipore Sigma, catalog number: M1891)
- [γ-32P]-ATP (Perkin Elmer, catalog number: NEG002Z), stored at -20 °C, Half-life is 14.29 days
- ATP (non-radioactive, Thermo Fisher Scientific, catalog number: PV3227)
- Phosphatase inhibitor (Sigma-Aldrich, catalog number: P0044)
- Magnesium chloride (MgCl2, Thermo Fisher Scientific, catalog number: AM9530G)
- Dithiothreitol (DTT, Thermo Fisher Scientific, catalog number: P2325)
- Ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA, Boston BioProducts, catalog number: BM-151)
- InstantBlue Coomassie Protein Stain (Expedeon, catalog number: ISB1L), stored at 4 °C
- Sodium dodecyl sulfate (SDS, Millipore Sigma, catalog number: 71725)
- SDS polyacrylamide gel (Precast or handcast gels can be used)
- Tris-HCl (Thermo Fisher Scientific, catalog number: 15567027)
- Beta-mercaptoethanol (Millipore Sigma, catalog number: M3148)
- Glycerol (Millipore Sigma, catalog number: G5516)
- Bromophenol blue (Millipore Sigma, catalog number: B3269)
- 10x kinase reaction buffer (see Recipes), stored at -20 °C in small aliquots
- 4x SDS sample buffer (see Recipes), stored at -20 °C in small aliquots
Equipment
- Pipettes
- -80 °C freezer
- Vertical mini-gel electrophoresis system (such as Mini-PROTEAN Tetra Vertical Electrophoresis Cell, Bio-Rad, catalog number: 1658005)
- Electrophoresis power supply (such as PowerPac Basic power supply, Bio-Rad, catalog number: 1645050)
- Perspex shielding and Perspex Eppendorf tube holders
- Geiger counter
- Heat blocks or water baths set to 30 °C and 95 °C
- Benchtop centrifuge
- Gel dryer (such as Bio-Rad gel dryer Model 583)
- X-ray film processor (such as the one from Konica, catalog number: SRX101A) placed in a dark room
- X-ray film cassette and security bag
- Scanner
Software
- ImageJ (National Institutes of Health and the Laboratory for Optical and Computational Instrumentation, USA, imagej.nih.gov/ij)
Procedure
This assay is performed by mixing ERK3 protein, purified substrate, 5 μCi [γ-32P]-ATP, and 30 μM non-radioactive ATP in a kinase reaction buffer that contains Mg2+. Fifty to one hundred nanograms of ERK3 protein purified from Sf9 cells or mammalian cells can be used per reaction. If using ERK3 protein purified from bacteria, a larger amount has to be used (500 ng-1 µg). It is best to do an initial optimization experiment using different amounts of the kinase to determine the kinase concentration appropriate for phosphorylating substrate. A catalytically-inactive (kinase-dead) mutant of ERK3 can serve as a negative control for the reaction. In addition, a reaction that lacks ERK3 would confirm the absence of contamination of the substrate with radioactive isotopes or co-purified kinases. As for all enzymatic assays, the amount of substrate used in each reaction has to be in excess so that it is not rate-limiting. Typically about 0.5-2 µg substrate is used per reaction.
After incubation of ERK3 with the substrate in the presence of ATP, the reaction is stopped by addition of sample buffer containing sodium dodecyl sulfate (SDS) followed by boiling. Reaction components are separated by gel electrophoresis. The gel is then stained with Coomassie blue solution, dried and visualized by autoradiography (Figure 1). 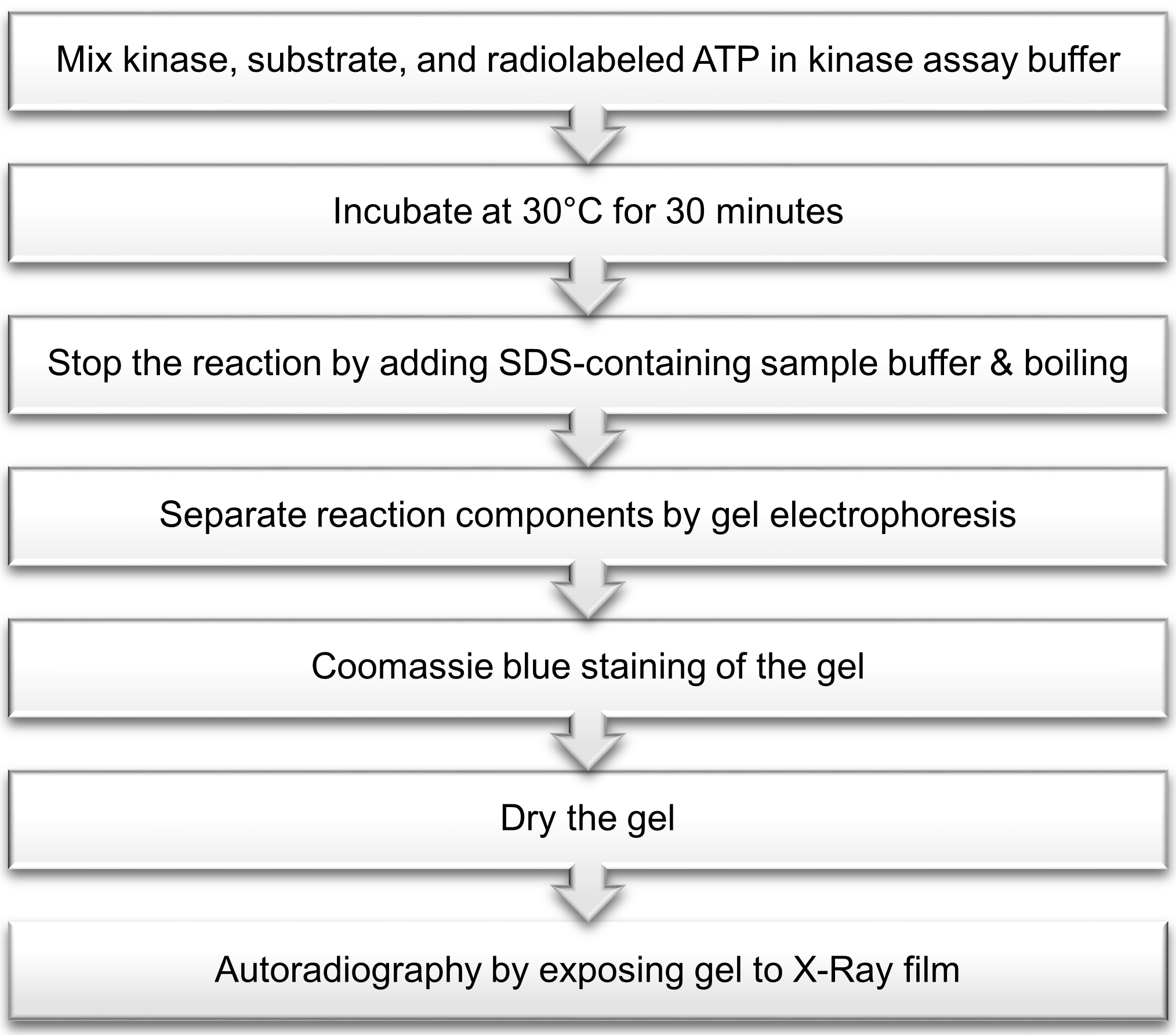
Figure 1. Outline of radioactive in vitro ERK3 kinase assay
Steps of the procedure are described below:
- Prepare a 10x kinase reaction buffer stock (listed in Recipes), and dilute an appropriate volume as needed with deionized water to 1x concentration.
- Dilute ERK3 protein and each purified substrate in 1x reaction buffer such that 1 μl of each is used per reaction. For example, if 100 ng ERK3 protein and 1 μg MBP will be used in the assay, dilute ERK3 to 100 ng/μl, and MBP to 1 μg/μl using 1x reaction buffer.
Note: It is recommended to store purified proteins at -80 °C in small aliquots to avoid multiple freeze-thaws. Thawing has to be done slowly on ice before an experiment is to be performed. - Prepare 0.5 mM non-radioactive ATP solution by diluting stock ATP in 1x reaction buffer. It is best to prepare fresh dilute ATP at the time of the experiment, and to avoid repeated freeze-thaws of the stock ATP.
- Determine the volume of each component to be used such that the total reaction volume is 30 μl (Table 1). The ATP mix added per reaction contains 5 μCi [γ-32P]-ATP and non-radioactive ATP that yields final concentration of 30 μM. For example, ATP mix for one reaction can be prepared by mixing 0.5 μl of radioactive ATP (10 μCi/μl) and 1.8 μl of 0.5 mM non-radioactive ATP.
Note: The rate of decay of [γ-32P]-ATP has to be taken into consideration when calculating the volume required per reaction. Manufacturers specify a calibration or reference date, which corresponds to the indicated activity of a radiolabeled reagent. This date can be used to determine the residual activity of the radioactive isotope on the day the assay is conducted. For example, for an experiment to be done two weeks after the [γ-32P]-ATP reference date, since the half-life of 32P isotopes is 14.29 days, double the volume of radiolabeled ATP has to be used to account for the decreased activity. Hence, instead of mixing 1.8 μl of 0.5 mM non-radioactive ATP with 0.5 μl of radioactive ATP as suggested above, 1.8 μl of 0.5 mM non-radioactive ATP should be mixed with 1 μl of radioactive ATP. The total volume of each reaction should always be adjusted to 30 µl.
Table 1. Components of each kinase reaction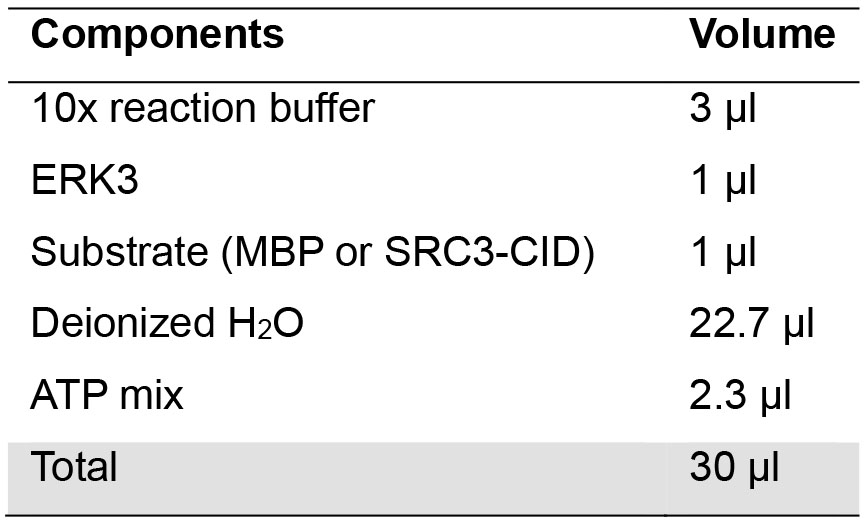
- Assemble reactions on ice by mixing all the components except for the ATP mix. If multiple reactions will use the same substrate, a master mix comprising 10x reaction buffer, substrate, and water can be prepared and distributed to the tubes to minimize pipetting errors.
- From this step onwards, precautions of using radioactive isotopes have to be taken. Prepare sufficient ATP mix for the number of assays to be performed. Start the reactions by adding an appropriate volume of ATP mix to each tube.
Note: Radioactive material handling precautions have to be taken. These include the use of appropriate shielding materials such as Perspex shielding (3/8 inches thick) behind which all work should be done, and Perspex Eppendorf tube holders. Surfaces should be routinely monitored by Geiger counters, and ring dosimeters can be used to monitor personal exposure. Radioactive isotopes should be used only by authorized personnel in designated places following the institution’s biosafety regulations. Requisition and storage of radioactive material, solid and liquid radioactive waste disposal, and spill decontamination should be done following the institution’s regulations. - Incubate the tubes in a water bath or sand bath at 30 °C for 30 min.
- Stop the reactions by adding 10 μl of 4x SDS sample buffer (listed in Recipes) followed by boiling at 95 °C for 5 min. Spin down the tubes to bring the reaction components to the bottom of the tube.
- Load the entire volume of each reaction into a well of a 10-well polyacrylamide gel. The percentage of resolving gel depends on the molecular weight of protein substrates used in the assay. Run the gel at 100-130 constant voltage. It is important to stop gel running when the dye front is about 1-2 cm away from the bottom of the gel to prevent the entry of free [γ-32P]-ATP into the buffer in the gel tank. Cut the gel just above the dye front and discard the lower portion into solid radioactive waste.
- Stain the gel with a sufficient volume of InstantBlue Coomassie protein stain for 30 min, followed by two washes using deionized water for 30 min each. Alternatively, standard Coomassie blue staining and destaining solutions can be used. Discard the solutions used for staining and washing into a liquid radioactive waste container.
- Place the gel on a thick filter paper, and dry it in a gel dryer at 70 °C for 60 min.
- Expose radioactivity with X-ray film for an appropriate time as follows:
- In a dark room place the gel in an X-ray film cassette with an X-ray film directly above it.
- Close the cassette and put it in an X-ray cassette security bag to ensure light protection. The use of intensifying screens enhances the signal. Also, keeping the cassette in a -80 °C freezer enhances the signal.
- Depending on the activity of the kinase towards the specific substrate (the phosphorylation level of the substrate), an X-ray film may be exposed for as short as 2-3 h, or as long as 1-2 days.
- Develop and fix the X-ray film to visualize substrate phosphorylation by autoradiography. Dark bands appearing on the X-ray film correspond to phosphorylated substrate that is isotope-labeled. Several exposures can be obtained to ensure that the signal is not over-saturated.
- A representative image of an in vitro ERK3 kinase assay using MBP as substrate is shown in Figure 2. The first lane is a negative control reaction which lacks ERK3. The second lane is a reaction that contains ERK3 and MBP. Note that ERK3 protein is barely seen in the Coomassie-stained gel because of its small amount, whereas the appearance of phosphorylated ERK3 band in the autoradiograph indicates ERK3 autophosphorylation.
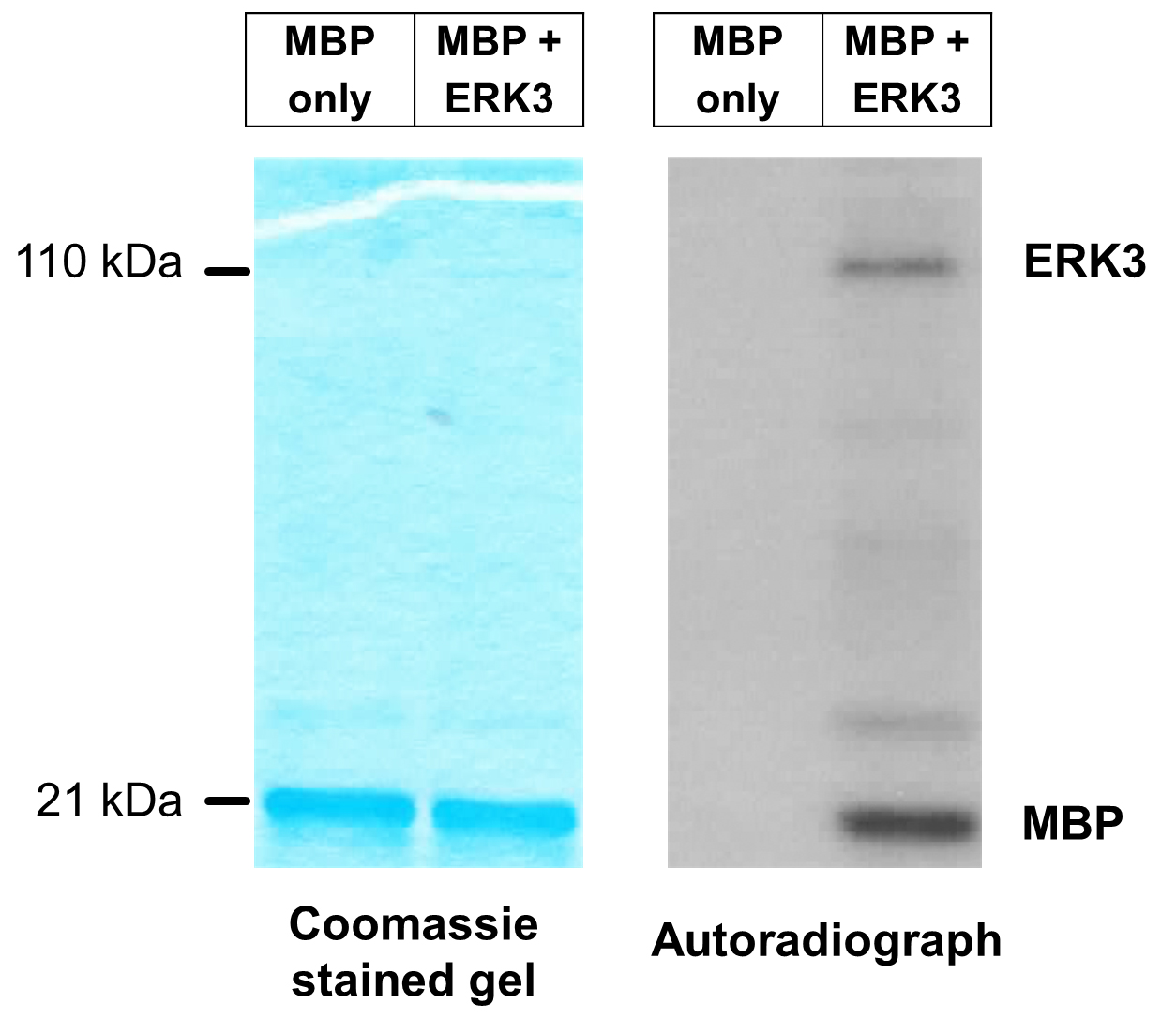
Figure 2. Representative image of in vitro ERK3 kinase assay using MBP as substrate. ERK3 protein with an N-terminal HA-tag (HA-ERK3) was expressed in 293T cells. HA-ERK3 protein was then purified by immunoprecipitation using HA antibody-conjugated beads, followed by elution with HA peptide. In vitro kinase reaction was performed in the presence of [γ-32P]-ATP and MBP (1 µg), with or without 100 ng purified ERK3 (lanes 2 and 1 respectively). Total protein levels of MBP in both reactions are shown by Coomassie staining (left panel). Phosphorylation of MBP is detected by autoradiography (right panel).
Data analysis
Data is analyzed by scanning the dry gel and the developed X-ray film using a standard scanner, and determination of the intensity of bands using image analysis software such as ImageJ. Quantification of substrate phosphorylation is done by calculating the ratio of the band intensity of phosphorylated substrate in the autoradiograph over that of the corresponding total substrate protein in the coomassie-stained gel. The ratio calculated for several samples can each be normalized to a reference condition. A hypothetical example to demonstrate data analysis is shown in Table 2:
Table 2. Demonstration of in vitro kinase assay data analysis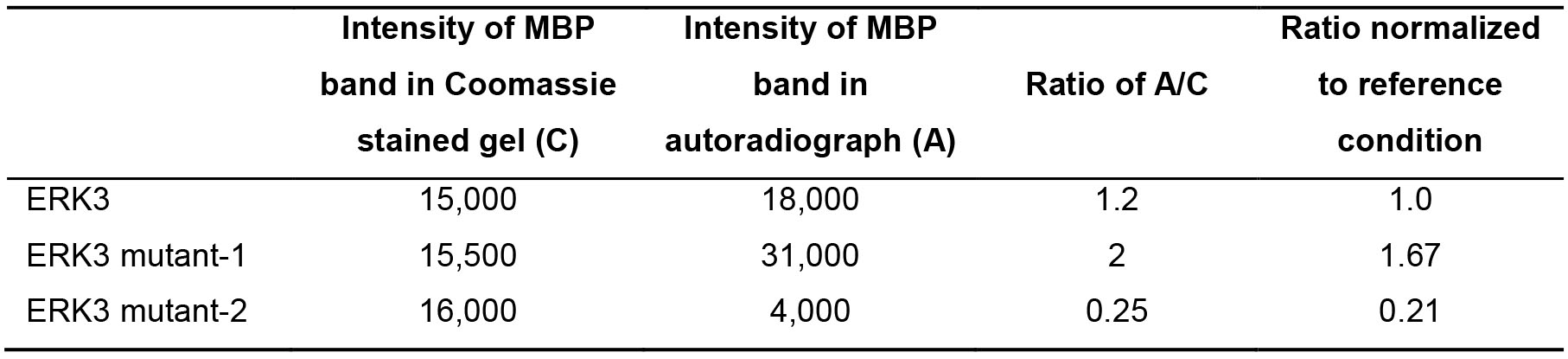
Recipes
- 10x kinase reaction buffer (1 ml)
Components Final concentration 400 μl 1 M Tris-HCl pH 7.5 400 mM Tris-HCl pH 7.5 100 μl 1 M MgCl2 100 mM MgCl2 10 μl 100 mM EGTA 1 mM EGTA 10 μl 1 M DTT 10 mM DTT 10 μl phosphatase inhibitor 1% Deionized water Add up to 1 ml - 4x SDS sample buffer (10 ml)
Components Final concentration 2.5 ml 1 M Tris-HCl pH 6.8 250 mM Tris-HCl pH 6.8 4 ml 100% glycerol 40% glycerol 0.8 g SDS 8% SDS 40 mg bromophenol blue 0.4% bromophenol blue 0.5 ml beta-mercaptoethanol 5% beta-mercaptoethanol Deionized water Add up to 10 ml
Acknowledgments
This protocol was modified from the research article by Cheng et al. (1996). This work was supported by a start-up fund of Wright State University and NCI 1R01CA193264-01 to Weiwen Long, and by the Biomedical Sciences PhD Program of Wright State University to Lobna Elkhadragy.
Competing interests
The authors declare no conflict of interest.
References
- Alsaran, H., Elkhadragy, L., Shakya, A. and Long, W. (2017). L290P/V mutations increase ERK3's cytoplasmic localization and migration/invasion-promoting capability in cancer cells. Sci Rep 7(1): 14979.
- Bian, K., Muppani, N. R., Elkhadragy, L., Wang, W., Zhang, C., Chen, T., Jung, S., Seternes, O. M. and Long, W. (2016). ERK3 regulates TDP2-mediated DNA damage response and chemoresistance in lung cancer cells. Oncotarget 7(6): 6665-6675.
- Cheng, M., Boulton, T. G. and Cobb, M. H. (1996). ERK3 is a constitutively nuclear protein kinase. J Biol Chem 271(15): 8951-8958.
- Coulombe, P. and Meloche, S. (2007). Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim Biophys Acta 1773(8): 1376-1387.
- Elkhadragy, L., Alsaran, H., Morel, M. and Long, W. (2018). Activation loop phosphorylation of ERK3 is important for its kinase activity and ability to promote lung cancer cell invasiveness. J Biol Chem 293(42): 16193-16205.
- Haubrich, B. A. and Swinney, D. C. (2016). Enzyme activity assays for protein kinases: strategies to identify active substrates. Curr Drug Discov Technol 13(1): 2-15.
- Long, W., Foulds, C. E., Qin, J., Liu, J., Ding, C., Lonard, D. M., Solis, L. M., Wistuba, II, Qin, J., Tsai, S. Y., Tsai, M. J. and O'Malley, B. W. (2012). ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion. J Clin Invest 122(5): 1869-1880.
- Schumacher, S., Laass, K., Kant, S., Shi, Y., Visel, A., Gruber, A. D., Kotlyarov, A. and Gaestel, M. (2004). Scaffolding by ERK3 regulates MK5 in development. EMBO J 23(24): 4770-4779.
- Seternes, O. M., Mikalsen, T., Johansen, B., Michaelsen, E., Armstrong, C. G., Morrice, N. A., Turgeon, B., Meloche, S., Moens, U. and Keyse, S. M. (2004). Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. EMBO J 23(24): 4780-4791.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Elkhadragy, L. and Long, W. (2019). A Radioactive in vitro ERK3 Kinase Assay. Bio-protocol 9(16): e3332. DOI: 10.21769/BioProtoc.3332.
- Elkhadragy, L., Alsaran, H., Morel, M. and Long, W. (2018). Activation loop phosphorylation of ERK3 is important for its kinase activity and ability to promote lung cancer cell invasiveness. J Biol Chem 293(42): 16193-16205.
Category
Biochemistry > Protein > Posttranslational modification
Molecular Biology > Protein > Phosphorylation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









