- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ex vivo Analysis of DNA Repair Capacity of Human Peripheral Blood Mononuclear Cells by a Modified Host Cell Reactivation Assay
Published: Vol 9, Iss 15, Aug 5, 2019 DOI: 10.21769/BioProtoc.3325 Views: 4625
Reviewed by: Lu SongAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for Quantifying γH2AX Foci in Irradiated Cells Using Immunofluorescence and Fiji Software
Lu Deng [...] Lingying Wu
Aug 20, 2025 2489 Views

Colocalizing Telomeres With PML or γH2AX Foci by IF-FISH in Mouse Brain Neurons
Anna Konopka
Nov 5, 2025 1531 Views

A Quantitative DNA Fiber Assay to Monitor Replication Fork Progression, Protection, and Restart
Debanjali Bhattacharya and Ganesh Nagaraju
Feb 5, 2026 292 Views
Abstract
The ability of humans to repair DNA damages decreases with increasing age. In order to be able to repair daily occurring DNA damages, it becomes more and more important to preserve repair capability of cells with aging. The preservation of DNA repair processes contributes to preventing DNA mutations and subsequently the onset of age-related diseases such as cancer. For the determination of DNA repair of human cells, mostly in vitro cell cultures are used. However, an ex vivo approach can provide a more accurate result compared with in vitro cell cultures, since the DNA repair ability is measured directly without the influence of prolonged culture time. Published protocols use in vitro cultured cells with a single reporter plasmid or a luciferase reporter. Our modified host cell reactivation assay enables the measurement of DNA repair capacity (nucleotide excision repair) of ex vivo isolated human peripheral blood mononuclear cells (PBMCs). For this purpose, PBMCs are isolated out of human anticoagulated blood by density gradient centrifugation. Directly after isolation, the PBMCs are co-transfected with two plasmids, one being previously damaged by UVC irradiation and one remaining undamaged. PBMCs are incubated for 24 h and subsequently analyzed by fluorescence activated cell sorting (FACS). The ability of cells to repair the DNA damages leads to a functional reactivation of the reporter gene. The assay presented here provides a solution to determine human DNA repair capacity ex vivo directly out of the human body. Furthermore, it can be used to research the ex vivo influence of different substances on DNA repair capacity of humans.
Keywords: Modified host cell reactivation assayBackground
Maintaining the integrity of the human genome is a prerequisite for delaying or preventing the development of age-related diseases. Every day, up to 50,000 DNA-damaging events occur in every cell (reviewed in Shrinivas et al., 2017). Such damages arise because of exogenous influences like radiation or chemicals, or via the normal cellular metabolism. For this reason, functional DNA repair mechanisms are crucial for the survival of organisms. One of the most important DNA repair pathways is nucleotide excision repair (NER), which detects and eliminates bulky DNA lesions and helix-distorting DNA adducts like cyclobutane pyrimidine dimers (CPDs) and pyrimidine-(6-4)-pyrimidone photoproducts (6-4PPs) (for review see Nouspikel, 2009).
The modified host cell reactivation assay (mHCRA) is a reliable and reproducible method for the research of NER capacity of human cells based on the restoration of a previously damaged reporter gene. The principle of reporter gene restoration was first described in 1985 (Protić-Sabljić et al., 1985), subsequently adjusted by Athas et al. for an application with human lymphocytes (Athas et al., 1991) and further refined by using fluorescent reporter proteins (Roguev and Russev, 2000). Up to now, few researchers use the mHCRA, although it is a highly reproducible and reliable method. Qiao et al. used the modified HCRA with a luciferase reporter on human lymphocytes to study DNA repair (Qiao et al., 2002). In 2010, Burger et al. published an advanced method, which describes the mHCRA for studies on human skin cells. They used two reporter plasmids and fluorescence-activated cell sorting to determine DNA repair capacity (Burger et al., 2010). Furthermore, Mendez et al. researched DNA repair by carrying out nucleofection of cryopreserved human lymphocytes with one reporter plasmid (Mendez et al., 2011).
Our mHCRA enables the measurement of NER capacity of PBMCs as an ex vivo approach. By using two different plasmids, transfection efficiencies can be normalized by a transfected control plasmid. This procedure allows for comparing experiments with different transfection efficiencies with each other. We used the method described here to reveal the influence of calorie reduction on the DNA repair capacity of humans (Matt et al., 2016). Moreover, the use of this assay can provide a more accurate assessment of positive or negative effects of substances like dietary supplements or pharmaceutical agents on DNA repair ability of human cells than the use of in vitro cell cultures.
Materials and Reagents
- Centrifuge tubes 15, 50 ml (SARSTEDT, catalog numbers: 62.554.001, 62.547.254)
- 1.5 ml reaction tube (SARSTEDT, catalog number: 72.706)
- Petri dishes (SARSTEDT, catalog number: 83.3902)
- Pipette tips 10 μl, 20 μl, 200 μl, 1,000 μl (SARSTEDT, catalog numbers: 70.1130, 70.116, 70.760.002, 70.762)
- Serological pipettes 5 ml and 10 ml (SARSTEDT, catalog numbers: 86.1253.001 and 86.1254.001)
- LeucosepTM, with porous barrier, 50 ml, pre filled with LeucosepTM separation medium (Greiner Bio-One GmbH, catalog number: 227288)
- Sterile filter 0.2 μm (SARSTEDT, catalog number: 83.1826.001)
- Cell culture plate, 6-well, Cell+ (SARSTEDT, catalog number: 83.3920.300)
- 0.4 cm electroporation cuvettes (Molecular BioProducts, Inc., purchased via Thermo Fisher Scientific GmbH, catalog number: 5540-11)
- FACS tubes 5 ml and 0.5 ml (SARSTEDT, catalog numbers: 55.1579, 55.673)
- Plasmid-DNA solution pEGFP-N1 (Clontech, catalog number: 6085-1) and pDsRedExpress-N1 (Clontech, catalog number: 632429), each 900 ng/µl in Qiagen EB buffer (storage temperature for longer periods: -80 °C, otherwise keep at 4 °C)
- Human venous blood, anticoagulated
- KCl (Carl Roth, catalog number: 6781.3)
- KH2PO4 (Carl Roth, catalog number: 3904.2)
- Na2HPO4·12H2O (Carl Roth, catalog number: N350.1)
- NaCl (Carl Roth, catalog number: 3957.3)
- S-Monovette® K3 EDTA 4.9 ml and 7.5 ml (SARSTEDT, catalog numbers: 04.1931.001 and 01.1605.001)
- RPMI 1640 without Phenol Red (Gibco® by Life Technologies, purchased via Fisher Scientific GmbH, catalog number: 11835-063, storage temp. 4 °C)
- Fetal Bovine Serum (Gibco® by Life Technologies, chased via Fisher Scientific GmbH, catalog number: 10270-106)
- EB Buffer, part of the QIAGEN Plasmid Plus Giga Kit (Qiagen GmbH, Hilden, catalog number: 12991)
- BD FACS Sheath Solution with Surfactant (BD Biosciences, catalog number: 336911)
- 1x PBS (see Recipes)
- 0.9% NaCl solution (see Recipes)
- PBMC medium (see Recipes)
Equipment
- Pipettes, Eppendorf Research® Plus 10 μl, 20 μl, 200 μl, 1,000 μl (Eppendorf, catalog numbers: 3123000020, 3123000039, 3123000055, 3123000063)
- Stratalinker® UV Crosslinker (Stratagene, catalog number: 400072, model: Model 1800)
- Centrifuge with swinging bucket rotor (Eppendorf, catalog numbers: 5805000010, 5804709004)
- Neubauer counting chamber improved (Carl Roth, catalog number: PC72.1)
- Gene Pulser II with Capacitance extender plus and Pulse controller plus (Bio-Rad Laboratories, Inc., catalog numbers: 165-2106, 165-2108, 165-2110)
- CO2 Incubator Hera cell 150 (Kendro Laboratory Products, catalog number: 51013568)
- BD FACSCalibur Flow Cytometry System with 488 nm laser (Becton, Dickinson and Company)
- -80 °C freezer HFU600TV (Thermo Fisher ScientificTM, catalog number: 11670823)
Software
- CellQuest Pro (Becton, Dickinson and Company, version 4.0.2)
- Prism (GraphPad Software, Inc.)
Procedure
Day 1
- Prepare plasmid solutions
- pDsRedExpress-N1: Adjust concentration to a maximum of 900 ng/µl using EB buffer.
Note: The higher the plasmid concentration is, the more likely plasmids lie above each other during irradiation and irradiation becomes less effective. This results in higher DNA repair capacities in the end. Irradiate half of the volume by pipetting droplets of 40 µl into a Petri dish (10 cm diameter). Irradiate with 5 kJ/m2 UVC (254 nm) without lid of the Petri dish using Stratalinker® UV Crosslinker. After irradiation, collect droplets in a 1.5 ml reaction tube. The other half of the plasmid solution remains non-irradiated. - pEGFP-N1 needs no further treatment as it serves as transfection control. The concentration of plasmid solution should be > 600 ng/µl in order to avoid too high volumes. The ratio of absorbance at 260 nm and 280 nm should be ~1.8.
- For the measurement of DNA repair, mix 10 µg of pEGFP-N1 and 30 µg of irradiated pDsRedExpress-N1. For calculation of the correction factor, mix 10 µg of pEGFP-N1 and 30 µg of non-irradiated pDsRedExpress-N1 for each electroporation. Note that the volumes needed depend on the concentration of the plasmid solutions. Adjust the volume of the mixture to 100 µl using EB buffer. Prepare enough plasmid-DNA mixtures for the entire study planned. Store mixtures at 4 °C for short-term storage or at -80 °C for longer periods.
- pDsRedExpress-N1: Adjust concentration to a maximum of 900 ng/µl using EB buffer.
- Isolate PBMCs from 12 ml of human blood using LeucosepTM tubes according to the manufacturer’s instructions as follows:
- Dilute 12 ml anticoagulated blood 1:2 using 0.9% NaCl solution (24 ml final volume) and transfer into LeucosepTM tubes.
- Centrifugate at 1,000 x g for 10 min in a swinging bucket rotor and switch off brakes. Figure 1A shows the LeucosepTM tube before centrifugation and Figure 1B shows the tube after centrifugation.

Figure 1. LeucosepTM tube before and after centrifugation. A. LeucosepTM tube filled with anticoagulated blood, which was previously diluted 1:2 with 0.9% NaCl solution. B. LeucosepTM tube after density gradient centrifugation. The different layer can be seen clearly from top to bottom: plasma, enriched cells, separation medium, the porous barrier, again separation medium and finally erythrocytes and granulocytes. - Pipet approximately 10 ml of plasma from the top in order to reach the enriched cell fraction more easily.
- Harvest the enriched cell fraction using a 1,000 µl Eppendorf pipette and transfer cells into a 50 ml centrifuge tube.
- Wash cells with 10 ml PBS and centrifuge at 250 x g for 10 min.
- Repeat the washing step two times each with 5 ml PBS. When adding 5 ml for the second time, count cells using a Neubauer counting chamber.
- For each electroporation, fill 3 ml PBMC medium per well into a 6-well cell culture plate.
- Centrifuge 2 x 106 cells at 250 x g for 10 min in a 15 ml centrifuge tube for each electroporation. Prepare two tubes in order to be able to calculate DNA repair capacity of one sample properly. One tube will contain the irradiated pDsRedExpress-N1 and pEGFP-N1 and the other one the non-irradiated pDsRedExpress-N1 and pEGFP-N1. Resuspend pellet in 150 µl RPMI media without phenol red.
- Add 100 µl of the previously prepared plasmid mixture to the cell suspension, so that the suspension contains 10 µg pEGFP-N1 and 30 µg pDsRedExpress-N1 (either irradiated or non-irradiated).
- Transfer 250 µl of the cell suspension to electroporation cuvettes with 4 mm gap, place it into the shocking chamber of Gene Pulser II and start electroporation at 400 V and 500 µF.
Note: Always electroporate one irradiated batch and one non-irradiated batch directly one after the other. These two will be used to calculate the DNA repair capacity of one sample. - Transfer electroporation batches in the order of electroporation into the prepared 6-well cell culture plate containing RPMI medium.
- Incubate for 24 h at 37 °C, 5 % CO2 and relative humidity of 95%.
- Prepare one 1.5 ml reaction tube for each electroporation approach.
- Mix cells in the wells of the cell culture plate using a 1,000 µl Eppendorf pipette before transferring 1.5 ml of the cell suspension into the 1.5 ml reaction tubes prepared previously.
- Centrifuge at 250 x g for 10 min.
- Remove 1.4 ml of the supernatant, resuspend cells in the remaining 100 µl, and transfer into 0.5 ml FACS tubes.
- Place the 0.5 ml tube into a 5 ml tube and carry out FACS analysis using 488 nm laser.
Data analysis
FACS analysis was carried out using FACSCalibur, CellQuest Pro software and the 488 nm laser. Gating of the cells of interest (lymphocytes) was carried out with the control sample. For analysis of the sample containing the irradiated pDsRedExpress-N1 the same gates were used. Gating of the transfected control samples and the samples containing the irradiated pDsRedExpress-N1 are shown in Figures 2 and 3 respectively.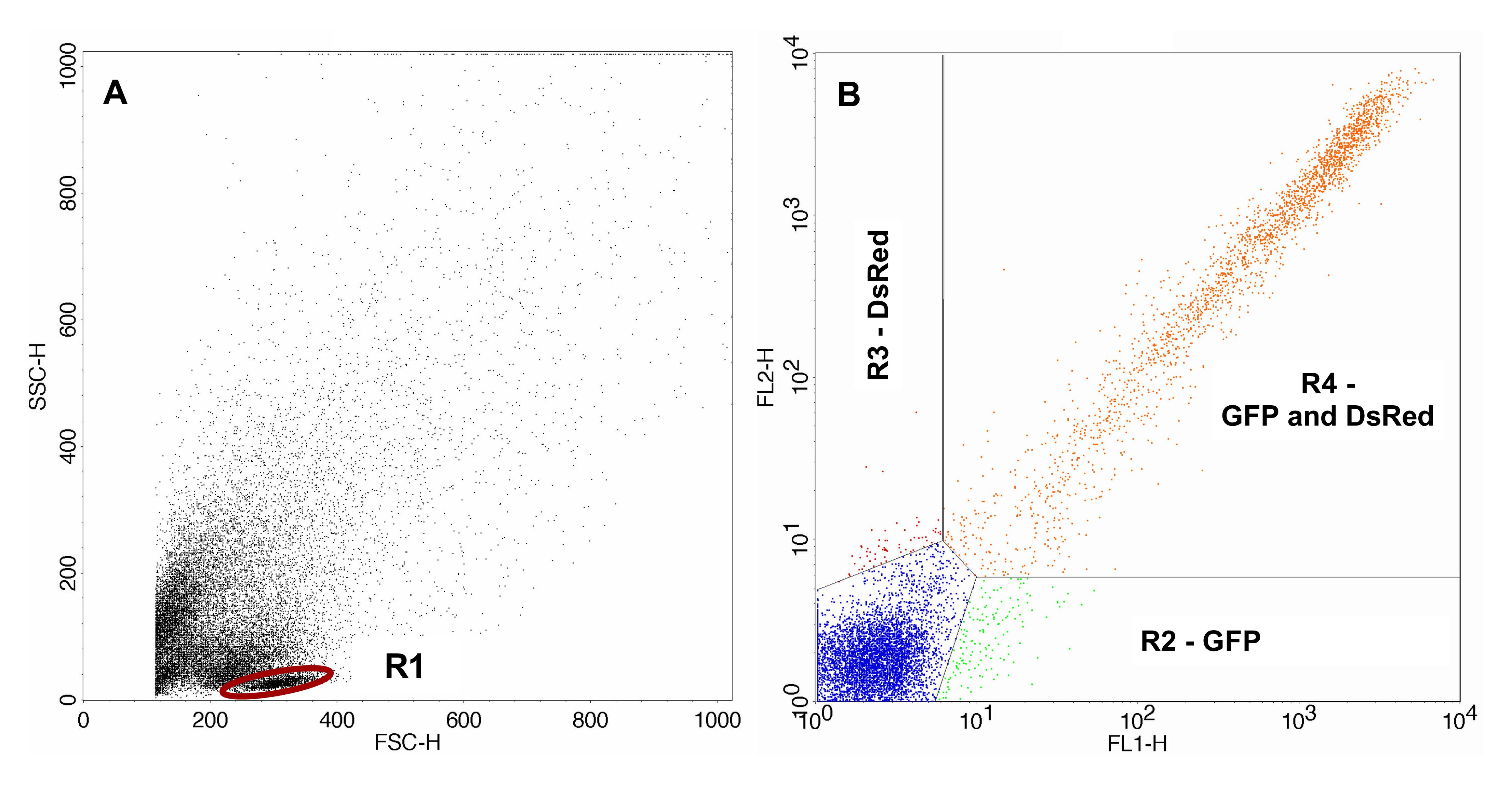
Figure 2. FACS analysis of the control sample. A. Control sample containing the non-irradiated pDsRedExpress-N1 with gated lymphocytes. B. Transfected cells gated with gate R1. The area lower right (R2) displays cells expressing only GFP, the area upper left (R3) displays cells expressing only DsRed and the area upper right (R4) displays cells expressing GFP as well as DsRed.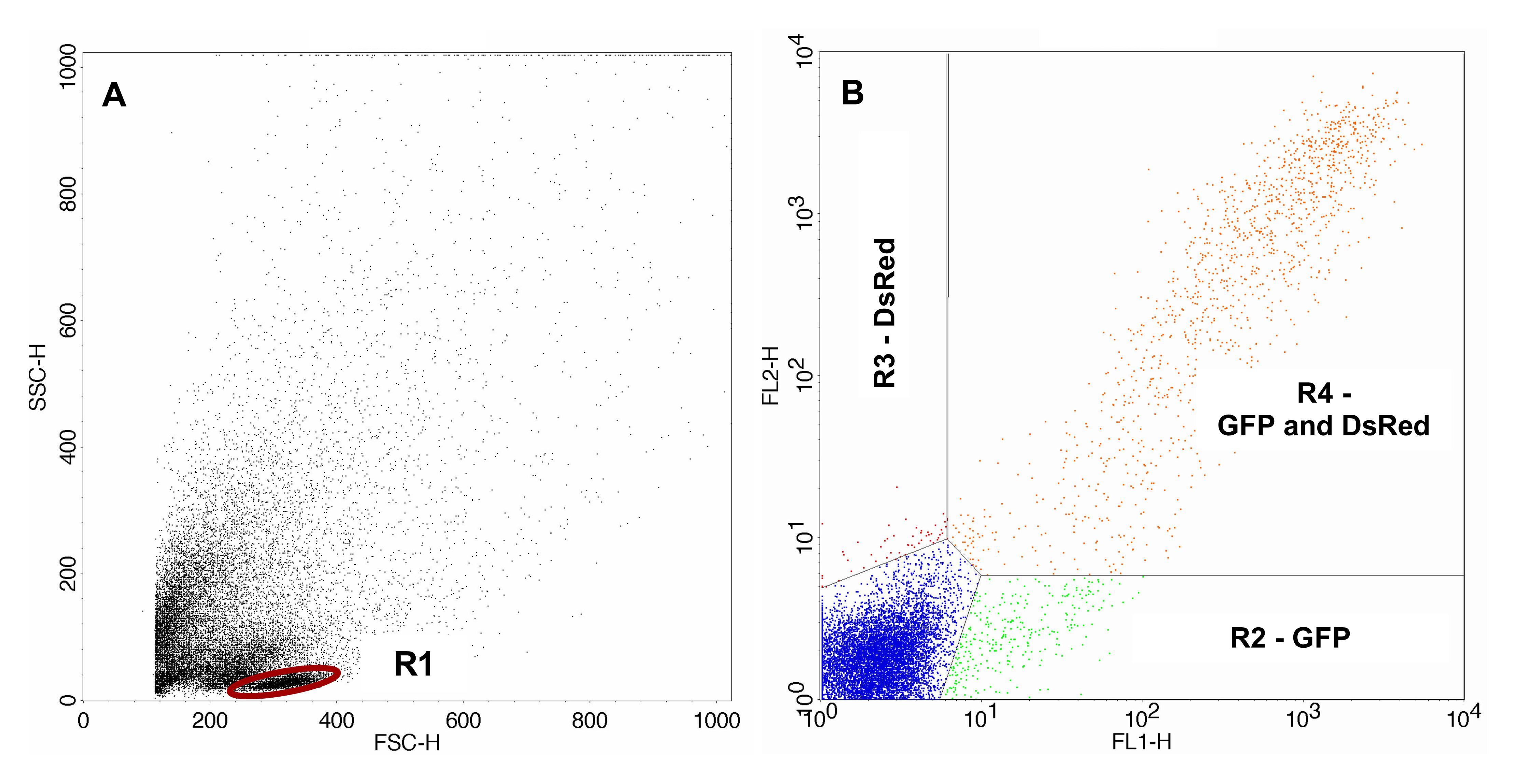
Figure 3. FACS analysis of the samples containing the irradiated pDsRedExpress-N1. A. Sample containing irradiated pDsRedExpress-N1 with gated lymphocytes. B. Transfected cells gated with gate R1. The area lower right (R2) displays cells expressing only GFP, the area upper left (R3) displays cells expressing only expressing DsRed and the area upper right (R4) displays cells expressing GFP as well as DsRed.
For calculation of DNA repair capacity, percentages of cells expressing GFP and DsRed are added up as follows. All GFP-expressing cells are the cells in gates R2 and R4. All DsRed-expressing cells are the cells in gates R3 and R4.
For the correction of repair capacity with regard to efficacy of transfection, the correction factor F is needed. F is always calculated from the percentages of GFP and DsRed of the control sample. Hence, a control is needed for each transfection with irradiated pDsRedExpress-N1. F is the ratio of percentages of GFP-expressing cells to the DsRed-expressing cells of the control sample. DNA repair capacity is calculated according to Matt et al. (2016) as follows:
For statistical analyses, any version of Prism (GraphPad Software, Inc.) can be used. We recommend triplicates for the calculation of DNA repair capacity.
Notes
- Pay attention to the time constants during electroporation. In our experiments, time constants are usually between 20 and 25 ms. Furthermore, too high time constants resulted in low transfection efficiencies or cell death.
- To allow the laser to warm up, switch on flow cytometer approximately 30 min before FACS analysis.
- To compensate for the non-optimal excitation of DsRed by the 488 nm laser, three times the amount of DsRed compared to GFP is used for electroporation.
- We recommend triplicates for each sample (three control approaches and three matching approaches containing the irradiated pDsRedExpress-N1).
Recipes
- 1x PBS
8 g NaCl
0.20 g KCl
2.88 g Na2HPO4·12H2O
1.24 g KH2PO4
Add 1 L ddH2O, pH 7.4 - 0.9% NaCl solution
9 g NaCl
Add 1 L ddH2O, sterile filter solution using 0.2 µm sterile filter - PBMC medium
RPMI 1640 without phenol red
20% fetal bovine serum
Store at 4 °C
Acknowledgments
The authors thank Dr. med. Karin Rupprecht (Center for Traditional Chinese Medicine–TCM Sigmaringen) and Dr. med. Adrian Schulte (F. X. Mayr Bodensee Center Überlingen) for drawing of the blood samples and informing of the volunteers.
Competing interests
One of the authors has consulting contracts with MSE Pharmazeutika GmbH, Bad Homburg, Germany.
Ethics
All experiments were conducted in accordance with the declaration of Helsinki as well as approved by the ethics committee of the Landesärztekammer Baden-Württemberg, Germany. The volunteers were informed before collection of the samples and gave their written consent to the use of their blood samples.
References
- Athas, W. F., Hedayati, M. A., Matanoski, G. M., Farmer, E. R. and Grossman, L. (1991). Development and field-test validation of an assay for DNA repair in circulating human lymphocytes. Cancer Res 51(21): 5786-5793.
- Burger, K., Matt, K., Kieser, N., Gebhard, D. and Bergemann, J. (2010). A modified fluorimetric host cell reactivation assay to determine the repair capacity of primary keratinocytes, melanocytes and fibroblasts. BMC Biotechnol 10: 46.
- Matt, K., Burger, K., Gebhard, D. and Bergemann, J. (2016). Influence of calorie reduction on DNA repair capacity of human peripheral blood mononuclear cells. Mech Ageing Dev 154: 24-29.
- Mendez, P., Taron, M., Moran, T., Fernandez, M. A., Requena, G. and Rosell, R. (2011). A modified host-cell reactivation assay to quantify DNA repair capacity in cryopreserved peripheral lymphocytes. DNA Repair (Amst) 10(6): 603-610.
- Nouspikel, T. (2009). DNA repair in mammalian cells-Nucleotide excision repair: variations on versatility. Cell Mol Life Sci 66(6): 994–1009.
- Protić-Sabljić, M., Whyte, D., Fagan, J., Howard, B. H., Gorman, C. M., Padmanabhan, R. and Kraemer, K. H. (1985). Quantification of expression of linked cloned genes in a simian virus 40-transformed xeroderma pigmentosum cell line. Mol Cell Biol 5(7): 1685-1693.
- Qiao, Y. W., Spitz, M. R., Guo, Z. Z., Hadeyati, M., Grossman, L., Kraemer, K. H. and Wei, Q. Y. (2002). Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutat Res 509(1-2): 165-174.
- Roguev, A. and Russev, G. (2000). Two-wavelength fluorescence assay for DNA repair. Anal Biochem 287(2): 313-318.
- Shrinivas, S. A., Shanta, S. H., Prajakta, B. B. (2017). DNA: damage and repair mechanisms in humans. Glob J Pharmaceu Sci 3(2): 555613.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Matt, K. C. and Bergemann, J. (2019). Ex vivo Analysis of DNA Repair Capacity of Human Peripheral Blood Mononuclear Cells by a Modified Host Cell Reactivation Assay. Bio-protocol 9(15): e3325. DOI: 10.21769/BioProtoc.3325.
Category
Molecular Biology > DNA > DNA damage and repair
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









