- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparation of cDNA Library for dRNA-seq
Published: Vol 2, Iss 23, Dec 5, 2012 DOI: 10.21769/BioProtoc.302 Views: 16015

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Mungbean Yellow Mosaic India Virus (MYMIV)-infection, Small RNA Library Construction and Deep Sequencing for MicroRNA Identification in Vigna mungo
Anirban Kundu [...] Genotypic Technology
Oct 20, 2016 11123 Views

Isolation of Nuclei in Tagged Cell Types (INTACT), RNA Extraction and Ribosomal RNA Degradation to Prepare Material for RNA-Seq
Mauricio A. Reynoso [...] Kaisa Kajala
Apr 5, 2018 16214 Views

In planta Transcriptome Analysis of Pseudomonas syringae
Tatsuya Nobori and Kenichi Tsuda
Sep 5, 2018 9349 Views
Abstract
microRNAs (miRNAs) are ubiquitous regulators of gene expression in eukaryotic organisms, which guide Argonaute proteins (AGO) to cleave target mRNA or inhibit its translation based on sequence complementarity. In plants, miRNA directed cleavage occurs on the target mRNA at about 10 to 11 nucleotide (nt) up stream to the site where the 5’ end of miRNA binds. Sequencing of the miRNA directed cleavage products (degradome) is widely employed as a way to both verify bioinformatic predictions of miRNA mediated regulation and identify novel targets of known miRNAs. Here we describe a protocol for preparation of degradome RNA complementary DNA library for high-through-put sequencing (dRNA-seq) using Illumina GA II sequencing platform, which is currently most popular and cost-effective. Using this protocol we successfully generated three dRNA-seq libraries using three solanaceae plants, including tobacco, tomato and potato. Although this protocol was developed with single-plexed adapter, it should be able to generate multiplexed libraries by replacing the 3’ adapter with multiplexing compatible 3’ adapter and replacing the PCR primer with indexed primers.
Keywords: NGSMaterials and Reagents
- RNeasy Plant Mini Kit (QIAGEN, catalog number: 74903 )
- OligodT Dynabeads (Life Technologies, Invitrogen™, catalog number: 610-02 )
- SeaKem LE Agrose (Lonza, catalog number: 50004 )
- Illumina sRNA-seq 3’ adapter (Illumina, catalog number: 1000596 )
- RNase free water (Life Technologies, Invitrogen™, catalog number: 10977-023 )
- RNeasy Micro Kit (QIAGEN, catalog number: 74004 )
- Antarctic phosphatase (New England Biolabs, catalog number: M0289S )
- RNase OUT(Life Technologies, Invitrogen™, catalog number: 10777-019 )
- T4 RNA Ligase 1 (New England Biolabs, catalog number: M0204S )
- Illumina sRNA-seq RT primer (Illumina, catalog number: 1000597 )
- Illumina sRNA-seq 5’ adapter (Illumina , catalog number: 1000595 )
- Illumina sRNA-seq PCR primer (Illumina, catalog number: 1000591 , 1000592 )
- Gel purification kit (QIAGEN, catalog number: 28704 )
- dNTP (New England Biolabs, catalog number: N0447S )
- SuperScript II RT(Life Technologies, Invitrogen™, catalog number: 18064-022 )
- Zero Blunt® PCR Cloning Kit (Life Technologies, Invitrogen™, catalog number: K2700-40 )
- Agrose gel (Lonza)
Equipment
- PCR Thermal Cycler
- Illumina GA II sequencing system
- Pipette (20 μl, 200 μl, 1,000 μl)
- Magnetic bar
Procedure
- Isolation of high molecular weight RNA (with length > 200 bp) from plant tissue using RNeasy Plant Mini Kit according to manufacturer’s protocol (according to the manufacturer’s protocol, about 60 μg high molecular weight RNA can be obtained from 100 mg tobacco leaf tissue).
- Purification of polyA RNA from 10 μg of total RNA using OligodT Dynabeads according to manufacturer’s protocol and elute the polyA RNA in 15 μl RNase free water (a thermal cycler and a magnetic bar are used in this step).
- Ligate sRNA 5’ adapter: 20 °C, 6 h.
Purified mRNA
14 μl
sRNA 5’ adapter (10 μM)
2 μl (Illumina sRNA-seq 5’ adapter)
10x T4 RNA Ligase buffer
2 μl (* If ATP is not included, add ATP to 1 mM final)
T4 RNA Ligase I (10 U/μl)
1.5 μl
RNase OUT (40 U/μl)
0.5 μl
- Dynabeads purification and elute in 15 μl RNase free water according to manufacturer’s protocol.
- RNA fragmentation 70 °C 2.5 min
Fragmentation buffer
1.6 μl (100 mM ZnCl2, 100 Mm Tris-HCl, pH7.0)
Ligated mRNA
14.4 μl
Purify fragmented RNA using RNeasy Micro Kit and elute RNA in 17 μl RNase free water after purification.
- Phosphotase treatment to remove 3’ phosphate resulted from fragmentation: 37 °C, 30 min
Fragmented RNA
16 μl
10x phosphatase buffer
2 μl
Antarctic phosphatase
1 μl
RNase OUT (40 U/μl)
1 μl
4 °C hold
Purify RNA by RNeasy Micro Kit and elute in 15 μl RNase free water.
- Ligate sRNA 3’ adaptor 20 °C, 4 h
Purified RNA from step 6
14.5 μl
10x RNA Ligase buffer
2 μl (* if ATP is not included, add ATP to 1 mM final)
RNA Ligase 1 (10 Uμl)
2 μl
RNase OUT (40 U/μl)
1 μl
RNA adapter 3’ 0.5 μl
(10 μM, Illumina sRNA-seq 3’ adapter)
- Reverse transcriptation
Prepare the following mix, heat at 70 °C for 2 min and place on ice.Prepare the following mix and add to the above reaction:Adapter ligated RNA
4 μl
SRA RT primer
0.5 μl (Illumina sRNA-seq RT primer)
50 mM dNTP
0.5 μl
48 °C for 3 min then add:5x first strand buffer
2 μl
100 mM DTT
2 μl
RNase OUT (40 U/μl)
0.25 μl
SuperScript II RT 0.75 μl
44 °C for 60 min
- PCR amplification
Prepare the following mix and add to RT reaction:
Run the following protocol:Phusion HF 2x mix
25 μl
Primer GX1
1 μl (Illumina sRNA-seq PCR primer)
Primer GX2
1 μl (Illumina sRNA-seq PCR primer)
Nuclease-free water
13 μl
- 98 °C 30 sec
- 30-35 cycles of:
98 °C 10 sec
60 °C 30 sec
72 °C 15 sec
- 72 °C 10 min
- 4 °C hold
- 98 °C 30 sec
- Run the PCR product through a 1.5% Agrose gel, cut a smear region between 150 bp and 250 bp and purify by Gel purification kit and elute in 25 μl elution buffer.
- Check the library quality by bio-analyzer High sensitivity DNA assay to check the size distribution (one μl sample is used in this step and a smear region centered around 250 bp is expected from the bio-analyzer electrophoresis profile, see Figure 1).
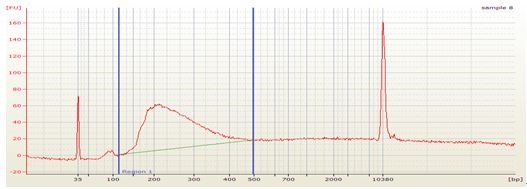
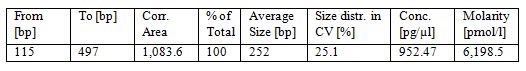
Figure 1. Bioanalyzer analysis of dRNA-seq library. The upper part is the electrophoresis graph from the bio-analyzer run and the peak region between the two blue lines represents the purified dRNA constructs. Table below the electrophoresis graph shows the analysis of the peak region by the bio-analyzer 2100 software.
- Use Zero Blunt® PCR Cloning Kit to clone the library and sequence of a few clones to verify the presence of inserts derived from plant transcripts.
- Sequence the library using small RNA sequencing run on an Illumina GA II sequencing system.
Acknowledgments
The principle and application of this protocol were briefly described in Li et al. (2012). This work was supported by the National Science Foundation Plant Genome Research Program Grant (DBI-0218166) and the United States Department of Agriculture (CRIS 5335-22000-007-00D). The authors declare no conflict of interest.
References
- Li, F., Orban, R. and Baker, B. (2012). SoMART: a web server for plant miRNA, tasiRNA and target gene analysis. Plant J 70(5): 891-901.
(Please cite this paper when you use this method in your publications)
Article Information
Copyright
© 2012 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Li, F. and Baker, B. (2012). Preparation of cDNA Library for dRNA-seq. Bio-protocol 2(23): e302. DOI: 10.21769/BioProtoc.302.
Category
Plant Science > Plant molecular biology > RNA > RNA sequencing
Molecular Biology > RNA > RNA interference
Systems Biology > Transcriptomics > RNA-seq
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










