- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Investigating Neural Stem Cell and Glioma Stem Cell Self-renewal Potential Using Extreme Limiting Dilution Analysis (ELDA)
(*contributed equally to this work) Published: Vol 8, Iss 17, Sep 5, 2018 DOI: 10.21769/BioProtoc.2991 Views: 11561
Reviewed by: Xi FengSubhra Prakash HuiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Melanoma Stem Cell Sphere Formation Assay
Alessandra Tuccitto [...] Michela Perego
Apr 20, 2017 15104 Views

Isolation and Culture of Neural Stem/Progenitor Cells from the Hippocampal Dentate Gyrus of Young Adult and Aged Rats
Mina Afhami [...] Koorosh Shahpasand
Oct 5, 2023 2271 Views

Derivation and Culture of Enriched Phrenic-Like Motor Neurons From Human iPSCs
Louise Thiry [...] Stefano Stifani
Jul 5, 2025 2290 Views
Abstract
Glioma stem cells (GSC) grown as neurospheres exhibit similar characteristics to neural stem cells (NSC) grown as neurospheres, including the ability to self-renew and differentiate. GSCs are thought to play a role in cancer initiation and progression. Self-renewal potential of GSCs is thought to reflect many characteristics associated with malignancy, including tumor recurrence following cytotoxic therapy due to their proliferative dormancy and capacity to allow for the development of resistant tumor cell sub-clones due to mutations acquired during their differentiation. Here, we demonstrate that using extreme limiting dilution analysis (ELDA), subtle differences in the frequency of sphere-forming potential between PI3K-mutant oncogenic NSCs and non-oncogenic NSCs can be measured, in vitro. We further show how ELDA can be used on cells, before and after forced differentiation to amplify inherent differences in sphere-forming potential between mutant and control NSCs. Ultimately, ELDA exploits a difference in the ability of a single or a few seeded stem cells to self-renew, divide and form neurospheres. Importantly, the assay also allows a comparison between genetically distinct cells or between the same cells under different conditions, where the impact of target-specific drugs or other novel cancer stem cell therapies can be tested.
Keywords: Stem cellsBackground
Glioblastoma (GBM) is one of the most common types of brain cancer with an extremely poor prognosis (Kaye and Morokoff, 2014). GBM treatment success or failure is determined by a complex biology due to the underlying genetics and epigenetics which regulate many signal transduction pathways, which in turn govern pro-oncogenic tumor cell behaviors and ultimately response/resistance to treatments. One of the major pathways regulating GBM cell behavior involves hyperactivation of PI3K signaling due mutations involving one or more pathway factors including the epidermal growth factor receptor (EGFR), the PI3K catalytic subunit encoded by the PIK3CA gene and deletion of the PI3K pathway negative regulator and tumor suppressor, Phosphatase and Tensin Homolog (PTEN) (Mantamadiotis, 2017). Using a genetically engineered mouse model, our recent study demonstrated that PI3K mutations, specifically targeting the Pik3ca and Pten genes in neural stem/progenitor cells (NSPs) can trigger the development of aggressive brain tumors, in vivo (Daniel et al., 2017 and 2018). In vitro analysis of NSCs isolated from mutant and wild-type mice revealed intrinsic cellular differences between genotypes, including sphere forming potential, which is a correlate of stem cell potential. This protocol describes the method used to investigate neural stem cell potential via established and variations on established methods. Extreme limiting dilution analysis (ELDA) can be used to analyze stem cell self-renewal ability and has been widely used in the stem cell field (Shackleton et al., 2006; Quintana et al., 2008; Vaillant et al., 2008). Subtle differences between NSCs or GSCs can be quantified by ELDA using a freely available webtool (Hu and Smyth, 2009). Moreover, we present a variation in stem cell potential analysis by including an intermediate differentiation step between ELDA measurements, which revealed more substantial differences in sphere forming potential between mutant and wild-type mouse NSCs, compared to performing ELDA alone, with cells which had not undergone forced differentiation. The forced differentiation step measures the inherent stability of NSCs and GBM-like NSCs, which maintain neurosphere forming potential following differentiation; stability which may correlate with faster disease relapse/recurrence following therapy.
Materials and Reagents
- Pipette tips: 10 ml, 5 ml, 2 ml (Axygen)
- 24-well tissue culture treated plate (Corning, catalog number: 3524 ) (Storage condition: 15 °C to 25 °C)
- 6-well tissue culture treated plate (Corning, catalog number: 3506 ) (Storage condition: 15 °C to 25 °C)
- 96-well Ultra-Low Attachment plate (Corning, catalog number: 3474 ) (Storage condition: 15 °C to 25 °C)
- 40 μm cell strainer (Corning, catalog number: 352340 ) (Storage condition: 15 °C to 25 °C)
- PIK3CAH1047R; PTENlox/lox; UBC-CreERT2 mice (Daniel et al., 2017 and 2018), aged E13.5 or 6 weeks old
Note: NSPCs can be isolated more efficiently from younger mice. Mice used here were on a C57BL/6 genetic background. - (Optional) Antibody
- anti-GFAP(glial fibrillary protein)
- anit-Tuj1
- DMEM F12 (Thermo Fisher Scientific, GibcoTM, catalog number: 10565042 ) (storage condition: 4 °C)
- Accutase cell detachment solution (BD, catalog number: 561527 ) (storage condition: 4 °C)
- Fetal Calf serum (FCS) (Bovogen Biologicals, catalog number: SFBS-FR ) (storage condition: -20 °C)
- Penicillin-streptomycin (10,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140-122 ) (storage condition: -20 °C)
- B-27 supplement (50x) (Thermo Fisher Scientific, GibcoTM, catalog number: 17504-044 ) (storage condition: -20 °C)
- Epidermal growth factor (EGF) (Thermo Fisher Scientific, GibcoTM, catalog number: PHG0313 ) (storage condition: -20 °C)
- Basic fibroblast growth factor (bFGF) (BioVision, catalog number: 4036 ) (storage condition: -20 °C)
- Laminin (Thermo Fisher Scientific, GibcoTM, catalog number: 23017-015 ) (storage condition: -20 °C)
- NaCl (Sodium Chloride, Thermo Fisher Scientific, Ajax Finechem, Univar, catalog number: AJA465-500G )
- KCl (Postassium Chloride, Merck, catalog number: 1049360500 )
- Na2HPO4 (di-Sodium hydrogen phosphate, Merck, catalog number: 106586 )
- KH2PO4 (Potassium dihydrogen orthophosphate, Thermo Fisher Scientific, Ajax Chemicals, catalog number: 392 )
- Phosphate buffered saline (PBS, see Recipes)
- Neurosphere culture media (see Recipes)
- Differentiated media (see Recipes)
Equipment
- Pipettes (METTLER TOLEDO, Pipet-X, model: PX-100 )
- Biohazard safety cabinet (EuroClone, model: BioAir-TopSafe 1.2-ABC )
- Centrifuge (Dynamica Scientific, model: Velocity 18R )
- DigiRetina16 camera (Tucsen)
- Neubauer Hemocytometer (Sigma-Aldrich, catalog number: Z359629 )
- Water bath at 37 °C (Froilabo, model: BMUTE)
- Humidified incubator at 37 °C, 5% CO2 (Thermo Fisher Scientific, Heraeus, model: HeracellTM 150 )
- Inverted phase contrast microscope (Nikon, model: Diaphot , with ELWD 0.3 phase contrast)
Software
- ELDA online software (http://bioinf.wehi.edu.au/software/elda/)
Procedure
- Neurosphere culture
Mutant and normal neural stem cells (NSCs) were isolated from a PIK3CA-PTEN mice, as described previously (Daniel et al., 2017 and 2018). This protocol can be used for both wild-type and mutant neurospheres grown in neurosphere culture media.- Dissociate NSCs to single cell suspension by incubating with Accutase cell detachment solution for 5 min at 37 °C.
- Stop the dissociation process by adding 2 ml of PBS.
- Filter the mixture through a 40 μm cell strainer.
- Add the neurosphere cell culture media to the filtrate.
- Centrifuge at 300 x g for 5 min.
- Resuspend single cell suspension in NSC culture media.
- Perform a cell count using a hemocytometer.
- Seed cells into a 24-well ultra-low attachment plate and allow sphere formation.
- Incubate plate in a 37 °C, 5% CO2 humidified incubator for 7 days.
- Replenish half the medium, every second day.
- Extreme limiting dilution assay (ELDA) for neurospheres
- Dissociate neurospheres into a single cell suspension, as described in Procedure A, Steps 1-6.
- Determine cell number, using a hemocytometer.
- Seed cells at decreasing densities (at 20, 10, 5, 1 cell/well, into a 96-well ultra-low attachment plate), in neurosphere culture media. Plate 24 replicates per cell density/number.
- Incubate in a 37 °C, 5% CO2 humidified incubator for 7 days (up to 14 days).
- From day three onward, monitor each well for signs of spheroid formation, using a phase contrast microscope; photograph, as necessary. Images shown were captured using a DigiRetina16 camera (http://www.tucsen.com).
- At seven days, score any wells that contain one or more sphere greater than ~20 μm diameter. Sphere size is measured using pre-calibrated scale bar function in the image capture software (TCapture5.1 software is free at http://www.tucsen.com).
- Input the data into ELDA software at http://bioinf.wehi.edu.au/software/elda/ in the format indicated in Data analysis, Step 3, below.
- Select “Plot Results” checkbox; default confidence interval is set at 0.95.
- Click “Run”.
- Results plot can be printed as a PDF and tabulated results can be used to generate a bar graph.
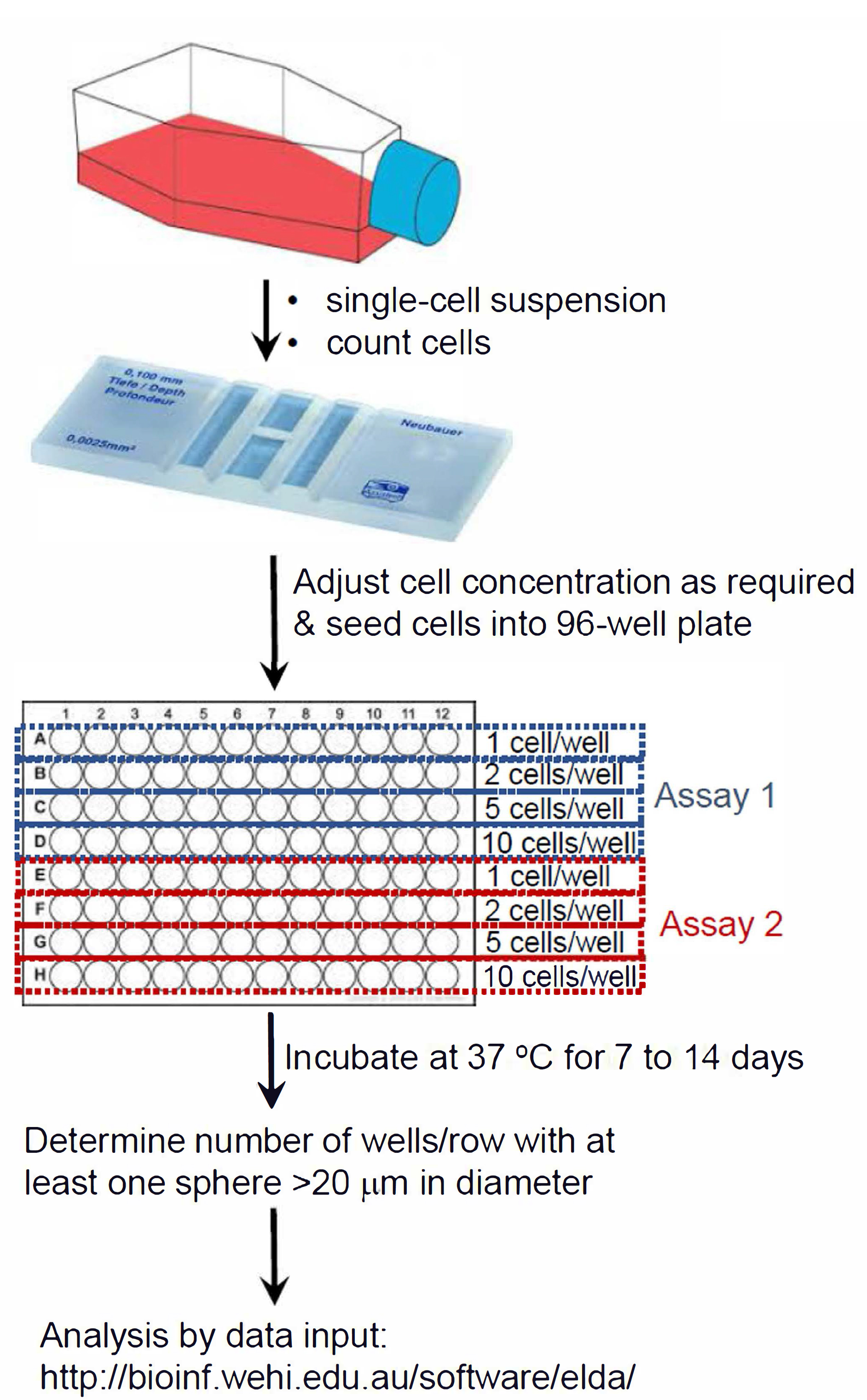
Figure 1. Summary schematic of ELDA setup
- Neural stem cell differentiation
This protocol is used to culture NSC cells in differentiation media for 7 days, harvest cells, then culture back in neurosphere medium (see Figure 2F).- Resuspend NSC single cell suspension in differentiation medium (DMEM-F12, 10% Fetal Calf Serum, penicillin-streptomycin [100 U/ml]).
- Seed 1 x 105 cells/well onto a 6-well standard adherent cell culture treated plate, pre-coated with laminin.
- Add 2 ml of differentiated medium to each well.
- Incubate in a 37 °C, 5% CO2 humidified incubator for 7 days to allow differentiation.
- Replenish medium every second day.
- Harvest cells at Day 7 and perform ELDA (Procedure B).
- Optionally, differentiation of NSCs can be checked in a separate cell preparation using anti-GFAP (glial fibrillary protein) antibody to mark glial cells, and anit-Tuj1 antibody to mark neuronal fated cells.
Data analysis
- Use a phase contrast microscope to monitor for sphere-forming cells and capture image of spheres between wild-type and mutant groups.
- Score the number of neurosphere-forming unit (NSFU)/number of cells seeded. Spheres > 20 μm in diameter were scored (Figures 2A-2C).
- Prepare an Excel spreadsheet with the following data.
- Input this information into the ELDA online software at http://bioinf.wehi.edu.au/software/elda/.
- Generate a plot with a 95% confidence interval to give sphere-forming frequency between wild-type or mutant groups (Figures 2D, 2E, 2G and 2H).
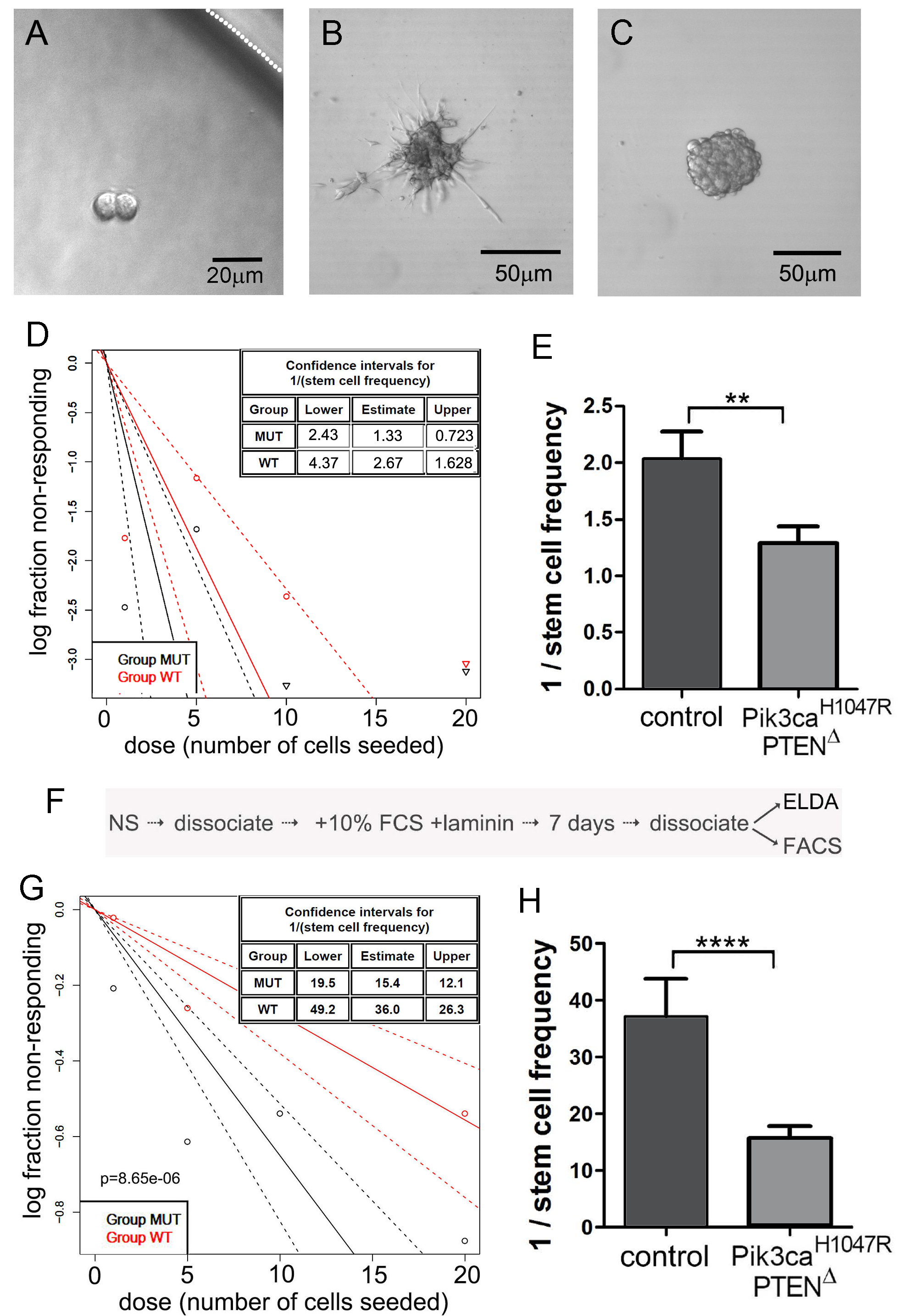
Figure 2. Extreme limiting dilution analysis (ELDA) for the assessment of stem-like cell self-renewal and a variation involving an intermediate step to differentiate NSCs. A. Representative NSCs following 7 days of culture after seeding a single NSC, showing two NSCs but no neurosphere formation. The white dotted line indicates the inner edge of a well on the 96-well plate. B. NSCs divided but spontaneous differentiated, showing no indication of neurosphere formation. C. A well showing formation of a single neurosphere after 7 days following seeding of a single NSC; this was scored as “positive”. D. Neurosphere forming capacity showing data output using the online algorithm (Procedure B). E. Graphical representation of the reciprocal of stem cell frequency calculated in (D). Note that PI3K mutant cells formed spheres more efficiently than control NSCs. F. The protocol flow after the initial ELDA, used to differentiate NSCs, then perform a second ELDA to measure inherent sphere forming stability. G and H. ELDA data output of the same NSC genotypes used for (D) and (E), following forced differentiation and replating in neurosphere medium.
Notes
ELDA assay can be used to study self-renewal ability of any type of cells. With ELDA software, the frequency of sphere-forming cells can be generated and compared between multiple cell sub-populations and different treatments.
Recipes
- 10x Phosphate buffered saline, pH 7.3
Milli-Q H2O
1,370 mM NaCl
27 mM KCl
81 mM Na2HPO4
14.7 mM KH2PO4 - Neurosphere culture media (final working concentrations)
DMEM/F12
Penicillin-streptomycin (100 U/ml)
1x B27 (from 50x stock)
10 ng/ml EGF
10 ng/ml bFGF - Differentiated media
DMEM/F12
Penicillin-streptomycin (100 U/ml)
10% FCS
Acknowledgments
This work was supported by the CASS Foundation Grant (#7491). A brief description of the protocol described in this article originally appeared in Daniel et al. (2017 and 2018). The original ELDA protocol is described in Hu and Smyth, 2009.
Competing interests
The authors declare no conflicts of interest.
References
- Daniel, P., Filiz, G., Brown, D., Christie, M., Waring, P., Zhang, Y., Haynes, J., Pouton, C., Flanagan, D., Vincan, E., Johns, T., Montgomery, K., Phillips, W. and Mantamadiotis, T. (2017). PI3K activation in neural stem cells drives tumorigenesis which can be suppressed by targeting CREB. BioRxiv 143388.
- Daniel, P. M., Filiz, G., Brown, D. V., Christie, M., Waring, P. M., Zhang, Y., Haynes, J. M., Pouton, C., Flanagan, D., Vincan, E., Johns, T. G., Montgomery, K., Phillips, W. A. and Mantamadiotis, T. (2018). PI3K activation in neural stem cells drives tumorigenesis which can be ameliorated by targeting the cAMP response element binding (CREB) protein. Neuro Oncol Apr 30. doi: 10.1093/neuonc/noy068.
- Hu, Y. and Smyth, G. K. (2009). ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347(1-2): 70-78.
- Kaye, A. H. and Morokoff, A. (2014). The continuing evolution: biology and treatment of brain tumors. Neurosurgery 61 Suppl 1: 100-104.
- Mantamadiotis, T. (2017). Towards targeting PI3K-dependent regulation of gene expression in brain cancer. Cancers 9(6): 60.
- Quintana, E., Shackleton, M., Sabel, M. S., Fullen, D. R., Johnson, T. M. and Morrison, S. J. (2008). Efficient tumour formation by single human melanoma cells. Nature 456(7222): 593-598.
- Shackleton, M., Vaillant, F., Simpson, K. J., Stingl, J., Smyth, G. K., Asselin-Labat, M. L., Wu, L., Lindeman, G. J. and Visvader, J. E. (2006). Generation of a functional mammary gland from a single stem cell. Nature 439(7072): 84-88.
- Vaillant, F., Asselin-Labat, M. L., Shackleton, M., Forrest, N. C., Lindeman, G. J. and Visvader, J. E. (2008). The mammary progenitor marker CD61/β3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res 68(19): 7711-7717.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Nguyen, H. P., Daniel, P. M., Filiz, G. and Mantamadiotis, T. (2018). Investigating Neural Stem Cell and Glioma Stem Cell Self-renewal Potential Using Extreme Limiting Dilution Analysis (ELDA). Bio-protocol 8(17): e2991. DOI: 10.21769/BioProtoc.2991.
Category
Stem Cell > Adult stem cell > Neural stem cell
Cancer Biology > Cancer stem cell > Cell biology assays > Self-renewal
Cell Biology > Cell-based analysis > Non-adherent culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link