- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Antisense Oligonucleotide-mediated Knockdown in Mammary Tumor Organoids
Published: Vol 7, Iss 16, Aug 20, 2017 DOI: 10.21769/BioProtoc.2511 Views: 11167
Reviewed by: Ralph BottcherAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Advances in Proximity Ligation in situ Hybridization (PLISH)
Monica Nagendran [...] Tushar J Desai
Nov 5, 2020 7366 Views

A Simplified 3D-Plasma Culture Method for Generating Minimally Manipulated Autologous Equine Muscle-Derived Progenitor Cells
Hélène Graide [...] Didier Serteyn
Dec 5, 2025 1279 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 212 Views
Abstract
Primary mammary tumor organoids grown in 3D are an excellent system to study tumor biology. They resemble the organization and physiology of native epithelia more closely than cancer cell lines grown in 2D, and additionally model interactions with the ECM (Boj et al., 2015; Clevers, 2016; Shamir and Ewald, 2014). Mammary tumor organoids are therefore a promising model system to identify and characterize novel drivers of breast cancer that would be unlikely to be identified using 2D cell lines. Antisense oligonucleotides can be used to efficiently and specifically knockdown target genes in the cell (Bennett et al., 2017). They can be taken up freely by organoids without the need for a transfection agent, making them a convenient tool for routine lab studies and screens.
Keywords: OrganoidsBackground
Breast cancer is the most frequent malignancy in women worldwide and the second leading cause of cancer mortality in women (Siegel et al., 2017). To improve existing treatment regimens, it is critical to identify and investigate new molecular targets that have the potential to prevent breast cancer progression. We applied RNA-seq to generate a comprehensive catalog of long non-coding RNAs (lncRNAs) that are dysregulated in primary mammary tumors compared to normal mammary epithelial cells and prioritized 30 previously uncharacterized lncRNAs as Mammary Tumor Associated RNAs (MaTARs). In order to functionally assess MaTARs as key drivers of tumor progression, we performed antisense oligonucleotide (ASO)-mediated knockdown assays of all 30 MaTARs in 3D mammary tumor organoids (Diermeier et al., 2016).
ASOs are short (20-mers), single stranded DNA molecules containing phosphorothioate-modified nucleotides as well as modifications of the 2’-ribose (5-10-5 2’-MOE gapmer) (Geary et al., 2015). Upon binding of the ASO to its complementary target, the RNA-DNA duplex stimulates degradation of the lncRNA by RNase H and thereby reduces the level of the respective transcript (Wu et al., 2004). Importantly, we found that ASO uptake in primary mammary tumor cells and organoids is efficient without the use of transfection agents, a mechanism that has been studied in detail in hepatocytes (Koller et al., 2011). ASO-mediated knockdown is particularly efficient for nuclear retained lncRNAs (Lennox and Behlke, 2016).
Organoids represent an ex vivo model of mammary gland development and model branching morphogenesis in 3D culture (Ewald, 2013; Fata et al., 2007), which is driven by two physiological processes: collective cell migration and cell proliferation. As the same processes also drive tumor invasion, the mammary organoid system can be utilized to model invasive breast cancer in vitro. Loss of branching was observed upon ASO-mediated knockdown of 20 MaTARs in organoids (Diermeier et al., 2016) as well as the lncRNA Malat1 (Arun et al., 2016), indicating that these RNAs are involved in mammary tumor cell proliferation and/or collective cell migration. Hence, we suggest that antisense-mediated knockdown in mammary tumor organoids can be used to identify and characterize novel drivers of tumor progression.
Materials and Reagents
- 5 cm sterile cell culture dish (e.g., Corning, Falcon®, catalog number: 353002 )
- Optional: Disposable plastic Cryomold (e.g., Tissue-Tek Cryomold, Electron Microscopy Sciences, catalog number: 62534-25 )
- 15 ml and 50 ml centrifuge tubes (e.g., Crystalgen, catalog number: 23-2265 ; Corning, Falcon®, catalog number: 352098 )
- 24-well cell culture plate (Greiner Bio One International, catalog number: 662160 )
- Cell culture flask, 75 cm2 (e.g., Corning, Falcon®, catalog number: 353136 )
- Pasteur glass pipettes (e.g., Kimble Chase Life Science and Research Products, catalog number: 63A54 )
- Sterile pipette tips (e.g., Corning)
- Cell strainer 70 micron (e.g., Corning, catalog number: 431751 )
- 0.2 ml PCR strip tubes (e.g., Corning, Axygen®, catalog number: PCR-0208-CP-C )
- 96-well reaction plates (e.g., Thermo Fischer Scientific, Applied BiosystemsTM, catalog number: 4346907 )
- Optical adhesive film (e.g., Thermo Fischer Scientific, Applied BiosystemsTM, catalog number: 4311971 )
- Mammary tumor-bearing mouse (e.g., MMTV-PyMT (Guy et al., 1992)) with palpable tumors. Optimal tumor size is ~5-10 mm in diameter
- 200 Proof ethyl alcohol (e.g., UltraPure, catalog number: 200CSPTP )
- Sterile water
- Ice
- Matrigel Growth Factor reduced Basement Membrane Matrix, phenol-red free (Corning, catalog number: 356231 )
- Optional: Tissue-Tek O.C.T. compound (Electron Microscopy Sciences, catalog number: 62550-12 )
- Liquid N2
- Cell recovery solution (Corning, catalog number: 354253 )
- 1x DPBS (e.g., Thermo Fisher Scientific, GibcoTM, catalog number: 14190250 )
- TRIzol (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15596018 )
- GlycoBlue (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9516 )
- DNAse I, amplification grade, for cDNA synthesis (e.g., Thermo Fisher Scientific, InvitrogenTM, catalog number: 18068015 )
- Ethylenediaminetetraacetate acid disodium salt (EDTA)
- Fetal bovine serum (FBS) (e.g., VWR, product number: 1500-500 )
- Gentamicin 50 mg/ml (e.g., Lonza, catalog number: 17-528Z )
- Insulin from bovine pancreas (Sigma-Aldrich, catalog number: I1882-100MG )
- Trypsin (e.g., Mediatech, catalog number: 25-054-CI )
- Collagenase from Clostridium histolyticum (Sigma-Aldrich, catalog number: C5138-1G )
- Advanced DMEM/F12 (e.g., Thermo Fisher Scientific, GibcoTM, catalog number: 12634010 )
- Bovine serum albumin (BSA) (e.g., Sigma-Aldrich, catalog number: A2153-100G )
- DNase I from bovine pancreas for organoid preparation (e.g., Sigma-Aldrich, catalog number: D4263-1VL )
- Pen/Strep (e.g., Sigma-Aldrich, catalog numbers: PENNA-100MU and S6501-100G )
- ITS liquid media supplement 100x (Sigma-Aldrich, catalog number: I3146-5ML )
- Murine FGF-basic (PeproTech, catalog number: 450-33 )
- Dimethyl sulfoxide (DMSO) (e.g., Sigma-Aldrich, catalog number: D2650-5x10ML )
- Nuclease-free water (e.g., Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9937 )
- TaqMan Reverse Transcription Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: N8080234 )
- PowerUp SYBR Green Master Mix (Thermo Fischer Scientific, Applied BiosystemsTM, catalog number: A25743 )
- Chloroform, purified (e.g., Avantor Performance Materials, MACRON, catalog number: 4432-10 )
- Isopropanol, molecular biology grade (e.g., Fisher Scientific, catalog number: BP2618500 )
- Collagenase solution (see Recipes)
- BSA solution (see Recipes)
- DNase solution (see Recipes)
- Organoid medium (10 ml, sufficient for 10 wells) (see Recipes)
- Freezing medium (see Recipes)
- cDNA Master mix (see Recipes)
- qPCR Master mix (see Recipes)
Equipment
- Biological safety cabinet (e.g., NuAire)
- Dissection tools (e.g., 114.3 mm scissors, Sklar Surgical Instruments, catalog number: 98-104 ; forceps, Sklar Surgical Instruments, catalog number: 97-751 , sterile scalpels, e.g., Sklar Surgical Instruments, catalog number: 06-3110 )
- Cell culture incubator (e.g., Heracell i Copper CO2 incubator, Thermo Fischer Scientific, Thermo ScientificTM, model: HeracellTM 150i and 240i , catalog number: 50116050)
- Shaker with temperature control (e.g., Thomas Scientific, catalog number: 1222U12)
Manufacturer: Benchmark Scientific, catalog number: H1000-M . - Centrifuge for 15 and 50 ml tubes (e.g., Eppendorf, model: 5804 )
- Centrifuge for 1.5 ml reaction tubes with cooling function (e.g., Eppendorf, model: 5427 R )
- Vacuum suction
- Micro-pipettes (e.g., Gilson, catalog number: F167300 , catalog number for Thomas Scientific: 1222N73)
- Phase-contrast microscope (e.g., Nikon)
- Optional: Cryo-Safe Freeze Controller (e.g., SP Scienceware - Bel-Art Products - H-B Instrument, catalog number: F18844-0000 )
- NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 2000 , catalog number: ND-2000)
- PCR machine (e.g., Applied Biosystems Proflex Thermocycler, Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4484073 )
- qPCR machine (e.g., Applied Biosystems StepOne Plus Real-Time PCR system, Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4376600 )
- -80 °C freezer (e.g., VWR, catalog number: 10160-728 )
- Water bath (e.g., PolyScience, catalog number: WB10A11B )
- Optional: liquid N2 freezers (e.g., VWR, catalog number: 82017-934 )
- Refrigerator (4 °C)
Procedure
- Design of antisense oligonucleotides (ASOs)
- Choose a target gene for knockdown. The protocol described here is specifically suitable for long non-coding RNAs, but knockdown of protein-coding transcripts is equally possible.
- Design 20-mers complementary to the RNA sequence. Usually, ASOs are designed complementary to exons, but in some cases ASOs targeting introns can be efficient as well. Design at least two specific ASOs (100% sequence identity) for the target sequence as well as a negative control (e.g., a scrambled ASO control, see (Diermeier et al., 2016)). It is recommended to start with up to ten individual ASOs for new targets, as not every ASO will result in an efficient knockdown.
- Use Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure the ASOs are specific to the intended target only. Stringency is important at this step–even one or two mismatches can result in unwanted off-target effects.
- ASOs used in knockdown experiments carry modified nucleotides to increase binding affinity. Of the 20 nucleotides, the first and last 5 nucleotides are modified with 2’-O-methoxy-ethyl (MOE). The 10 nucleotides in the middle remain unmodified. In the US, MOE modifications can be ordered e.g., from Integrated DNA Technologies (IDT).
- Upon arrival, resuspend lyophilized ASOs in sterile DPBS. Working stock concentration: 200 µM.
- Store at 4 °C for short-term storage (up to one week) and -20 °C for long-term storage. Aliquot to avoid repeated freeze-thaw cycles.
- Choose a target gene for knockdown. The protocol described here is specifically suitable for long non-coding RNAs, but knockdown of protein-coding transcripts is equally possible.
- Preparation of mouse mammary tumor organoids
- Euthanize mammary tumor-bearing mouse according to the local regulations.
- Pin mouse with abdominal side up to dissection board, spray down mouse with 70% ethanol and transfer to sterile cell culture hood. All steps from here on are to be performed under sterile conditions.
- Remove tumor using sterile dissection tools and place it in a 5 cm cell culture dish. In case of multiple tumors, prepare organoids from each tumor separately.
- Remove necrotic tissue from tumor if present using a scalpel. In general, necrotic tissue is darker and softer compared to the surrounding solid tumor. Necrotic tissue can often be found as dark liquefied tissue at the center of large tumors (Morton and Houghton, 2007).
- Optional: It is recommended to store a small part of the tumor for histological analysis. Therefore, use a scalpel and carefully cut about ¼ of the tumor. Embed the tumor in O.C.T. in a plastic Cryomold and store at -80 °C.
- Cut the remaining tumor tissue with a scalpel until the tissue pieces are about 2-3 mm in diameter.
- Transfer the tumor tissue to a 50 ml tube containing 25 ml of collagenase solution (Recipe 1).
- Shake the collagenase solution containing tumor tissue in an incubator at 100 rpm at 37 °C for 45 min. The suspension now appears cloudy. Over-digestion leads to a large amount of single cells, which should be avoided.
- Pre-warm DMEM/F12 medium and organoid medium to RT.
- Pre-warm a 24-well cell culture plate (see Notes 1 and 2) and a 75 cm2 cell culture flask filled with sterile water to 37 °C.
- Thaw Matrigel on ice.
- Centrifuge tube with collagenase solution at 520 x g for 10 min at 25 °C.
- Aspirate the supernatant using a Pasteur glass pipette connected to vacuum suction, leaving about 1 ml of medium on top of the pellet.
- Pre-coat a 10 ml sterile pipette tip by pipetting BSA solution (Recipe 2) up and down once.
Important: All plastic pipettes and tubes should be pre-coated with BSA solution immediately before use from this point on to avoid sticking of organoids to the plastic surface, unless otherwise specified. - Resuspend the pellet in 10 ml of pre-warmed DMEM/F12 medium using the pre-coated pipette tip.
- Transfer the suspension to a 15 ml centrifugation tube that has been pre-coated with BSA solution.
- Centrifuge tube at 520 x g for 10 min at 25 °C.
- Aspirate the supernatant as described in step B13. Pasteur pipettes need not be pre-coated with BSA before use.
- Resuspend the pellet in 4 ml of DNase solution (Recipe 3) using a pre-coated 10 ml pipette tip.
- Gently rock by hand for 2-5 min.
- Add 6 ml of DMEM/F12 and resuspend thoroughly with a pre-coated 10 ml pipette tip.
- Filter the suspension through a cell strainer into a fresh, pre-coated 15 ml tube to remove large chunks that remain after collagenase digestion.
- Centrifuge tube at 520 x g for 10 min at 25 °C.
- Aspirate the supernatant as described in step B13. Pasteur pipettes need not be pre-coated with BSA before use.
- Resuspend the pellet in 10 ml of DMEM/F12 medium using a pre-coated pipette tip.
- Pulse-centrifuge tube to 520 x g at 25 °C. Stop the centrifuge as soon as it reaches 520 x g.
- Repeat steps B25-B27 three more times (for a total of four times) or until the medium after puls-centrifugation is clear. At the last aspiration step, carefully remove the entire medium.
- Resuspend the pellet in 10 ml of DMEM/F12 medium using a pre-coated pipette tip.
- Make sure the suspension is well mixed. Remove 50 µl using a micropipette and transfer to a sterile 5 cm cell culture dish to determine the density of organoids in solution.
- Count the number of organoids in the 50 µl drop under a microscope. Organoids are counted in the dish, no cover slips are required. An example of a mammary tumor organoid right after preparation is shown in Figure 1. Depending on the size of the tumor, the total amount of organoids per preparation can vary from approximately 5,000 to 40,000. Organoids that are not needed for the current experiment can be frozen and stored as described in step B42.
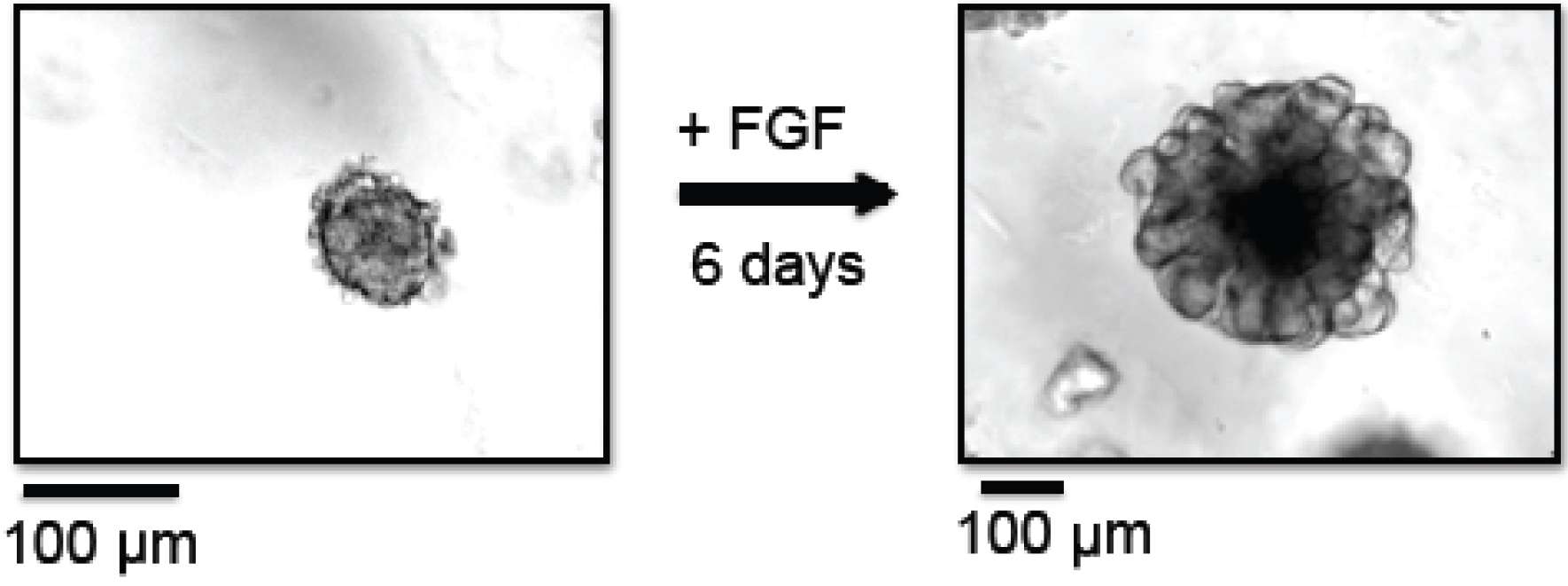
Figure 1. Branching morphogenesis of organoids. In this example, organoids were prepared from MMTV-PyMT derived mammary tumors. Left panel: Organoids at day 0, right after organoid preparation, embedded in Matrigel. Right panel: Organoids at day 6, grown in the presence of FGF-basic, embedded in Matrigel. - Centrifuge tube at 520 x g for 2 min at 25 °C.
- Aspirate the supernatant completely and carefully using a Pasteur glass pipette. Pasteur pipettes need not be pre-coated with BSA before use.
- Resuspend the organoid pellet in Matrigel based on the following criteria and put on ice immediately. Pipette tips and cell culture plates need not be pre-coated with BSA for this step due to the high protein content of Matrigel. To plate organoids in 24-well cell culture plates, 80 µl are required per well. Organoid plating density varies depending on the downstream assay. For microscopy analysis, plate organoids at a density of 2 organoids/µl (see Note 2). For RNA extractions, plate organoids at a density of 5 organoids/µl. One well yields a sufficient amount of RNA to perform qRT-PCR assays downstream; if higher RNA yields are required, pool two or more wells when extracting RNA (see Procedure C: RNA extraction from organoids).
- Prepare a sterile, pre-warmed (37 °C) 24-well cell culture plate and place it on top of a pre-warmed (37 °C) 75 cm2 cell culture flask filled with sterile water. The water-filled flask acts like a heated plate and is crucial for quick solidification of Matrigel.
- Carefully plate the Matrigel containing organoids as 80 µl domes in the middle of the wells (see Note 1). Avoid air bubbles. Pipette tips need not be pre-coated with BSA for this step.
- Incubate the plates in a cell culture incubator at 5% CO2 and 37 °C for 20 min.
- Add 1 ml of pre-warmed (RT) organoid medium (Recipe 4) per well. Be careful not to disturb the Matrigel domes.
- Add antisense oligonucleotides (ASOs) to the medium under sterile conditions. ASOs are added individually, one per well. Optimal ASO concentrations vary depending on the ASO and the target gene. A titration in the range from 0.1-5 µM should be performed (see Note 3). As a starting point, a concentration of 5 µM per well (25 µl of a 200 µM ASO solution) should result in a > 50% knockdown for most targets. ASOs are taken up freely by mammary organoids, no transfection reagent is required.
- Leave at least one well untreated and treat at least one well with a scrambled ASO (scASO) as negative controls.
- Incubate organoids in a cell culture incubator at 5% CO2 and 37 °C.
- Change the medium to fresh organoid medium and replenish ASOs three days past organoid generation. Organoids should be harvested no later than 6 days post generation (see Note 4). An example of a mammary organoid cultured for 6 days in the presence of FGF is shown in Figure 1.
- Optional: Any extra organoids that are not needed for the current experiment can be frozen and stored. To do so, calculate the amount of required organoids and respective volume of the organoid suspension at step B30. Split the organoid suspension in two aliquots, one containing the amount needed for the experiment and the other containing any additional organoids. Proceed with both aliquots as described in steps B31 and B32. Resuspend the organoids needed for the experiment in Matrigel and proceed as described above. Resuspend the extra organoids in freezing medium (Recipe 5), aliquot in cryovials and transfer to a CryoSafe freeze controller. Store at -80 °C overnight and transfer to liquid N2 the next day. To thaw organoids, warm the vial to 37 °C and resuspend the organoids in 10 ml of DMEM. Follow steps B31 to B37 to plate the organoids in Matrigel. For thawed organoids, a seeding density of 5-10 organoids/µl is recommended, as not all organoids will survive the freeze/thaw process.
- Euthanize mammary tumor-bearing mouse according to the local regulations.
- RNA extraction from organoids
- Organoids can be harvested as early as 24 h post ASO addition to assay the knockdown efficiency. If phenotypic analysis is required as well, live-cell imaging can be performed from day 3-day 6 after ASO addition (the day of organoid preparation is day 0), as the branching morphogenesis starts around day 3-4. Alternatively, endpoint microscopic analysis can be performed on day 6 to compare ASO treated organoids to control organoids. In this case, knockdown efficiency can still be assayed on day 6 (see Note 4).
- Transfer the 24-well plate containing organoids from the cell culture incubator to the refrigerator (4 °C). This step will cause the Matrigel to liquefy overnight.
- The next day, transfer the content of each well (medium and Matrigel) to a 15 ml centrifugation tube on ice.
- Add 200 µl of cold cell recovery solution to each well to depolymerize any residual Matrigel and incubate for 1 h at 4 °C.
- Transfer the content of each well (cell recovery solution and Matrigel) and combine with the previously removed organoid/Matrigel mix. If Matrigel sticks to the bottom, careful scraping may be used to detach Matrigel and organoids.
- Incubate on ice for 1 h.
- Centrifuge at 420 x g for 10 min at 4 °C.
- Aspirate the supernatant carefully, using a micropipette.
- Resuspend the organoid pellet in 100 µl of cold cell recovery solution.
- Incubate on ice for 10 min.
- Centrifuge at 420 x g for 5 min at 4 °C.
- Aspirate the supernatant carefully, using a micropipette.
- Resuspend the pellet in 1 ml of 1x DPBS (RNase-free).
- Centrifuge at 420 x g for 5 min at 4 °C.
- Aspirate the supernatant carefully, using a micropipette.
- Resuspend the cell pellet in 1 ml TRIzol.
- Isolate RNA from organoids according to the manufacturer’s instructions. Addition of glycogen (such as GlycoBlue) is recommended, as the RNA yield can be low if organoids were seeded at low density and harvested after 24 h. Resuspend RNA pellet in 10-20 µl of nuclease-free water.
- Organoids can be harvested as early as 24 h post ASO addition to assay the knockdown efficiency. If phenotypic analysis is required as well, live-cell imaging can be performed from day 3-day 6 after ASO addition (the day of organoid preparation is day 0), as the branching morphogenesis starts around day 3-4. Alternatively, endpoint microscopic analysis can be performed on day 6 to compare ASO treated organoids to control organoids. In this case, knockdown efficiency can still be assayed on day 6 (see Note 4).
- cDNA synthesis and qPCR to analyze knockdown efficiency
- Measure the RNA concentration using a NanoDrop 2000 UV-Vis spectrophotometer. Keep RNA on ice at all times to avoid degradation.
- Use a total of 1,000 ng RNA in a volume of 8 µl (concentrations of 125 ng RNA/µl). If concentrations are higher, dilute in nuclease-free water.
- Transfer sample to 0.2 ml PCR tubes.
- Perform DNase digestion to remove any potentially co-purified genomic DNA by adding 1 µl of 10x DNase reaction buffer and 1 µl DNase I to the RNA sample (total volume: 10 µl).
- Mix reaction by gently pipetting up and down. Spin briefly in a centrifuge.
- Incubate reaction at 25 °C in a PCR machine for 15 min.
- Add 1 µl of 25 mM EDTA.
- Mix reaction by gently pipetting up and down. Spin briefly in a centrifuge.
- Inactivate reaction at 65 °C in a PCR cycler for 10 min.
- Transfer samples on ice immediately after the 10 min incubation is completed. Do not leave samples in PCR machine while ramping down to RT.
- Prepare cDNA Master mix (Recipe 6).
- Add 39 µl of cDNA Master mix to samples on ice.
- Mix reaction by gently pipetting up and down. Spin briefly in a centrifuge.
- Perform the following incubation in a PCR machine:
25 °C for 10 min
48 °C for 30 min
95 °C for 5 min - cDNA can be stored short-term at 4 °C or long-term at -20 °C.
- Prepare qPCR Master mix (Recipe 7) containing primers for the target gene (see Note 5) as well as qPCR Master mix containing primers for an internal control (‘housekeeping’) gene. When calculating the amount of Master mix needed, consider that samples will be pipetted in triplicates.
- Commonly used genes for internal controls include beta-actin, GAPDH or ribosomal protein genes. Internal controls should be chosen carefully; their expression level should be consistent and independent on experimental conditions such as ASO treatments. The expression level of an internal control gene should be in the same range as the target gene. General rules regarding qPCR experiments and qPCR primer design can be found in (Bustin et al., 2009).
- Prepare a 96-well plate on ice.
- Pipette 2 µl of cDNA into one well of the 96-well plate. Each sample will be pipetted in triplicates for both the target and the housekeeping gene (= 6 wells containing 2 µl of cDNA per sample).
- Add 18 µl of Master mix containing either target gene or internal control gene primers to the wells.
- Seal the 96-well plate with an optical adhesive film. Avoid touching the film with your fingers, handle carefully by the edges.
- Spin down the plate at 100 x g for 1 min at RT.
- Place the plate in the qPCR machine and run the following program:
95 °C for 10 min
40 cycles of: 95 °C for 15 sec
60 °C for 60 sec
Followed by
Melt curve 95 °C → 60 °C, 1 °C/min
- Measure the RNA concentration using a NanoDrop 2000 UV-Vis spectrophotometer. Keep RNA on ice at all times to avoid degradation.
Data analysis
- Export Ct values from the qPCR machine and analyze using the 2-ΔΔCt Method (Livak and Schmittgen, 2001). There should be a noticeable reduction of the target gene in samples treated with ASOs compared to untreated samples and samples treated with scASO as negative control. The scASO should not have any effect on gene expression and result in values very similar to untreated samples. Very potent ASOs can result in a > 90% knockdown efficiency, but knockdown efficiencies vary between different ASOs and target genes. At least three independent biological replicates should be performed for statistically meaningful results. Insufficient knockdown efficiencies can be improved by increasing the amount of ASOs used, testing ASOs targeting different regions of the RNA and varying organoid densities. In addition, ASOs can be mixed with Matrigel when plating the organoids and also added to the medium for maximum knockdown efficiency.
- Endpoint analysis of phenotypic changes in organoids can be performed using standard light microcopy. When comparing ASO-treated organoids to control organoids, changes in the number, size and branching morphogenesis are often observed. To perform statistically significant comparisons, at least 100 organoids per treatment should be counted per replicate, with a minimum of three experimental replicates.
Notes
- Organoids can also be grown in 48-well and 96-well plates for assays requiring higher throughput. The size for the Matrigel domes has to be scaled down accordingly, e.g., 40 µl/well in 48-well plates and 20 µl/well in 96-well plates.
- If live-cell imaging is to be performed to analyze organoids, use glass-bottom plates (e.g., MatTek)
- If using new, untested ASOs, it is recommended to test several concentrations (e.g., 0.1, 0.5, 1 and 5 µM) and compare the knockdown efficiency.
- According to our RNA-seq data, culturing of organoids in an artificial extracellular matrix (ECM) for up to seven days does not significantly alter the expression patterns of lncRNAs (Diermeier et al., 2016).
- For optimal results, primers for qPCR should not overlap with the ASO binding site, and the ASO should not bind within the qPCR amplified region.
Recipes
- Collagenase solution (50 ml, sufficient for two tumors)
2 ml FBS
2.5 µl gentamycin (50 mg/ml)
25 µl insulin
4 ml trypsin
75 mg collagenase
Add DMEM/F12 up to 50 ml
Sterile filter before use
Use immediately after preparation - BSA solution (2.5%)
Dissolve 2.5 mg BSA in 100 ml DPBS
Sterile filter before use
Can be re-used and stored at 4 °C for up to four weeks - DNase solution
Add 40 µl of DNase I from bovine pancreas (1 U/µl) to 10 ml of DMEM/F12 medium
Use immediately after preparation - Organoid medium (10 ml, sufficient for 10 wells)
100 µl Pen/strep
100 µl ITS
3 µl FGF-basic
Add DMEM/F12 up to 10 ml
Can be stored at 4 °C for one week
Without addition of FGF2, organoid medium can be stored at 4 °C for up to four weeks - Freezing medium
10% DMSO in FBS
Can be stored long-term at -20 °C - cDNA Master mix (all components except for nuclease-free H2O are included in the TaqMan Reverse Transcription Kit)
Use immediately after preparation10x TaqMan RT buffer 5 µl Random hexamers 2.5 µl MgCl2 (25 mM) 11 µl dNTPs 10 µl RNase Inhibitor 1 µl RTase 1.25 µl Nuclease-free H2O 8.25 µl Total volume 39 µl - qPCR Master mix
Use immediately after preparationPowerUp SYBR Green Master mix 10 µl Nuclease-free H2O 7 µl Total 17 µl Primer mix (for + rev, 10 µM) 1/well (conc.: 0.5 µM/well)
Acknowledgments
Our tumor organoid protocol is based on previous work from Andrew Ewald’s lab (Ewald, 2013; Nguyen-Ngoc et al., 2015). The Manhasset Women’s Coalition Against Breast Cancer (S.D.D.) and the NCI 5P01CA013106-Project 3 (D.L.S.) supported this research.
References
- Arun, G., Diermeier, S., Akerman, M., Chang, K. C., Wilkinson, J. E., Hearn, S., Kim, Y., MacLeod, A. R., Krainer, A. R., Norton, L., Brogi, E., Egeblad, M. and Spector, D. L. (2016). Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 30(1): 34-51.
- Bennett, C. F., Baker, B. F., Pham, N., Swayze, E. and Geary, R. S. (2017). Pharmacology of antisense drugs. Annu Rev Pharmacol Toxicol 57: 81-105.
- Boj, S. F., Hwang, C. I., Baker, L. A., Chio, II, Engle, D. D., Corbo, V., Jager, M., Ponz-Sarvise, M., Tiriac, H., Spector, M. S., Gracanin, A., Oni, T., Yu, K. H., van Boxtel, R., Huch, M., Rivera, K. D., Wilson, J. P., Feigin, M. E., Ohlund, D., Handly-Santana, A., Ardito-Abraham, C. M., Ludwig, M., Elyada, E., Alagesan, B., Biffi, G., Yordanov, G. N., Delcuze, B., Creighton, B., Wright, K., Park, Y., Morsink, F. H., Molenaar, I. Q., Borel Rinkes, I. H., Cuppen, E., Hao, Y., Jin, Y., Nijman, I. J., Iacobuzio-Donahue, C., Leach, S. D., Pappin, D. J., Hammell, M., Klimstra, D. S., Basturk, O., Hruban, R. H., Offerhaus, G. J., Vries, R. G., Clevers, H. and Tuveson, D. A. (2015). Organoid models of human and mouse ductal pancreatic cancer. Cell 160(1-2): 324-338.
- Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., Mueller, R., Nolan, T., Pfaffl, M. W., Shipley, G. L., Vandesompele, J. and Wittwer, C. T. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4): 611-622.
- Clevers, H. (2016). Modeling development and disease with organoids. Cell 165(7): 1586-1597.
- Diermeier, S. D., Chang, K. C., Freier, S. M., Song, J., El Demerdash, O., Krasnitz, A., Rigo, F., Bennett, C. F. and Spector, D. L. (2016). Mammary tumor-associated RNAs impact tumor cell proliferation, invasion, and migration. Cell Rep 17(1): 261-274.
- Ewald, A. J. (2013). Isolation of mouse mammary organoids for long-term time-lapse imaging. Cold Spring Harb Protoc 2013(2): 130–133.
- Fata, J. E., Mori, H., Ewald, A. J., Zhang, H., Yao, E., Werb, Z. and Bissell, M. J. (2007). The MAPKERK-1,2 pathway integrates distinct and antagonistic signals from TGFα and FGF7 in morphogenesis of mouse mammary epithelium. Dev Biol 306(1): 193-207.
- Geary, R. S., Norris, D., Yu, R. and Bennett, C. F. (2015). Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 87: 46-51.
- Guy, C. T., Cardiff, R. D. and Muller, W. J. (1992). Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 12(3): 954-961.
- Koller, E., Vincent, T. M., Chappell, A., De, S., Manoharan, M. and Bennett, C. F. (2011). Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res 39(11): 4795-4807.
- Lennox, K. A. and Behlke, M. A. (2016). Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res 44(2): 863-877.
- Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4): 402-408.
- Morton, C. L. and Houghton, P. J. (2007). Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc 2(2): 247-250.
- Nguyen-Ngoc, K. V., Shamir, E. R., Huebner, R. J., Beck, J. N., Cheung, K. J. and Ewald, A. J. (2015). 3D culture assays of murine mammary branching morphogenesis and epithelial invasion. Methods Mol Biol 1189: 135-162.
- Shamir, E. R. and Ewald, A. J. (2014). Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol 15(10): 647-664.
- Siegel, R. L., Miller, K. D. and Jemal, A. (2017). Cancer Statistics, 2017. CA: a Cancer Journal for Clinicians 67(1): 7-30.
- Wu, H., Lima, W. F., Zhang, H., Fan, A., Sun, H. and Crooke, S. T. (2004). Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J Biol Chem 279(17): 17181-17189.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Diermeier, S. and Spector, D. L. (2017). Antisense Oligonucleotide-mediated Knockdown in Mammary Tumor Organoids. Bio-protocol 7(16): e2511. DOI: 10.21769/BioProtoc.2511.
Category
Cancer Biology > General technique > Molecular biology technique > Antisense oligonucleotide
Molecular Biology > RNA > qRT-PCR
Cell Biology > Cell isolation and culture > 3D cell culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link