- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Oxidative Stress Assays (arsenite and tBHP) in Caenorhabditis elegans
Published: Vol 7, Iss 13, Jul 5, 2017 DOI: 10.21769/BioProtoc.2365 Views: 12847
Reviewed by: Peichuan ZhangTugsan TezilMichael Enos

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Evaluation of Burkholderia cepacia Complex Bacteria Pathogenicity Using Caenorhabditis elegans
Pietro Tedesco [...] Donatella de Pascale
Oct 20, 2016 7690 Views

Artificial Optogenetic TRN Stimulation of C. elegans
Ithai Rabinowitch [...] Jihong Bai
Oct 20, 2016 8610 Views
Aerotaxis Assay in Caenorhabditis elegans to Study Behavioral Plasticity
Qiaochu Li [...] Karl Emanuel Busch
Aug 20, 2022 2238 Views
Abstract
Cells and organisms face constant exposure to reactive oxygen species (ROS), either from the environment or as a by-product from internal metabolic processes. To prevent cellular damage from ROS, cells have evolved detoxification mechanisms. The activation of these detoxification mechanisms and their downstream responses represent an overlapping defense response that can be tailored to different sources of ROS to adequately adapt and protect cells. In this protocol, we describe how to measure the sensitivity to oxidative stress from two different sources, arsenite and tBHP, using the nematode C. elegans.
Keywords: Hydrogen peroxideBackground
Reactive oxygen species (ROS) are small molecules that can damage DNA, proteins, lipids and other cellular components. Systemic levels of ROS induce irreversible cellular damage, which has been implicated in the etiology of aging and age-related diseases, such as Alzheimer’s disease, atherosclerosis, and diabetes. Furthermore, environmental toxins such as pollutants, smoke, chemicals, radiation, and xenobiotics significantly induce ROS formation. To protect against oxidative damage, cells have evolved complex mechanisms that detoxify ROS. Interestingly, long-lived animals show an enhancement of these protective mechanisms, implicating their importance for healthy aging.
The multicellular organism C. elegans has been instrumental in elucidating the molecular mechanisms that protect against ROS (Blackwell et al., 2015). In C. elegans, the major ROS detoxification mechanisms are initiated by the transcription factor SKN-1, the orthologue of the Nrf (nuclear factor-erythroid-related factor) proteins (Blackwell et al., 2015). Exposing C. elegans to either the metalloid sodium arsenite (As) or tert-Butyl hydroperoxide (tBHP; an organic peroxide) activates SKN-1, which promotes survival. Although overlapping sets of genes are upregulated by SKN-1 in response to As or tBHP, there are also condition-specific gene sets that tailor the oxidative stress response (Oliveira et al., 2009). Moreover, the expression of almost all detoxification genes in response to As depends on SKN-1, whereas the induction of several genes upon tBHP-treatment is also independent of SKN-1 (Oliveira et al., 2009), suggesting the activation of other oxidative stress response transcription factors. How different ROS sources are sensed and integrated is not well understood, but recently a mechanism has been elucidated for how As-induced ROS are generated and sensed by the cell (Hourihan et al., 2016).
Treating C. elegans with As activates the BLI-3/NADPH oxidase complex to produce localized pools of ROS, which modify a cysteine in the IRE-1 kinase and induce the SKN-1-dependent antioxidant response leading to lifespan extension (Hourihan et al., 2016). Further supporting the function of localized ROS levels as cell signals, recent work has identified a novel regulator of aging, MEMO-1, which increases resistance to As toxicity and facilitates lifespan extension in a BLI-3- and SKN-1-dependent manner (Ewald et al., 2017).
Taken together, oxidative stress responses can be induced directly, by exogenously added ROS sources such as tBHP, or as a secondary response to a chemical (such as As) or other stress leading to increased ROS levels. These two ROS sources elicit common, but also distinct downstream stress-response genes and protection mechanisms. Here, we describe the protocols for both ROS sources to assess C. elegans survival under these oxidative stress conditions.
Part I. Protocol for As stress tolerance assay
Materials and Reagents
- Pipette tips
- 24-well plates (Multiwell Culture Plate 24 well fl. NUNC, clear)
- C. elegans strains (available at Caenorhabditis Genetics Center [CGC] https://cbs.umn.edu/cgc/home)
- Nematode growth medium (NGM) culturing C. elegans plates (He, 2011) seeded with bacteria (OP50) grown overnight (300 μl OP50 per plate)
- M9 buffer (Physiological buffer for C. elegans; [He, 2011])
- Sodium arsenite solution volumetric, 0.05 M NaAsO2 (Honeywell International, catalog number: 35000 )
- Agar (He, 2011)
- Phosphate buffer (He, 2011)
- Calcium chloride (CaCl2) (He, 2011)
- Magnesium sulfate (MgSO4) (He, 2011)
- Cholesterol (He, 2011)
Equipment
- Pipettes
- Stereomicroscope
- Worm pick
Procedure
- Day 0 (Monday): Using the worm pick (Figure 1), transfer larval 4 stage (L4) worms onto fresh NGM culturing plate (He, 2011). Store these worms overnight at 20 °C.
Note: You need about 50 L4 worms per strain; e.g., 10-12 for each of the triplicates with As and for the control condition (M9 buffer only).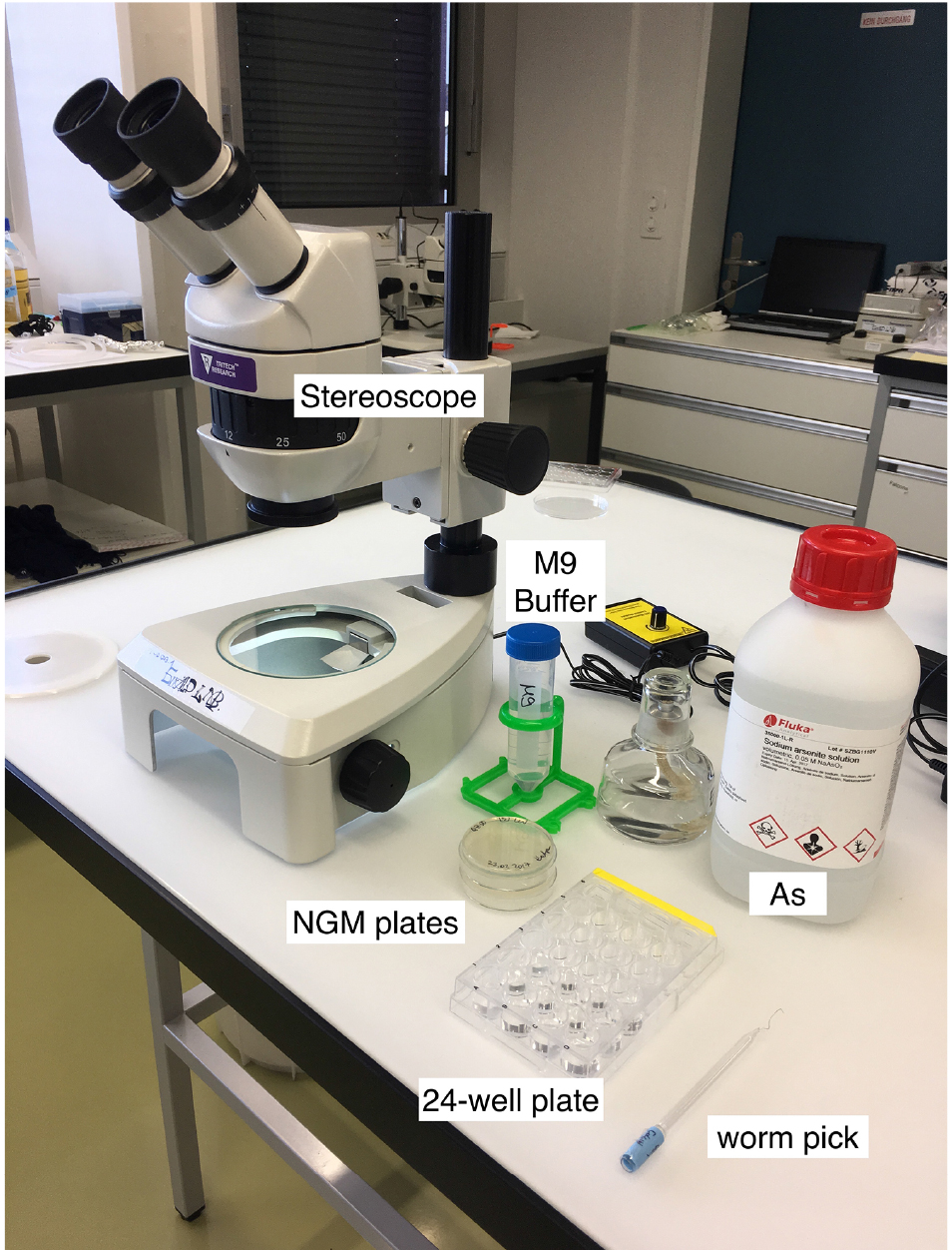
Figure 1. Material for the As-survival assay - Day 1 (Tuesday): Put 50 μl of M9 buffer into each well of a 24-well plate. See Figure 2 for loading scheme. Place 10-12 one-day-old worms into the M9 (Figure 3 and Video 1). You need 3 wells per strain for As and 1 well for M9 buffer as a control. When all worms are set, fill wells with either 450 μl of 5.56 mM As (for a final concentration of 5 mM As dissolved in M9) or M9.
- Score every hour for worm survival. (Exploded animals need to be excluded from the statistics.)
Important: The 5 mM As in M9 dilution must be prepared fresh directly before you put it into the wells.
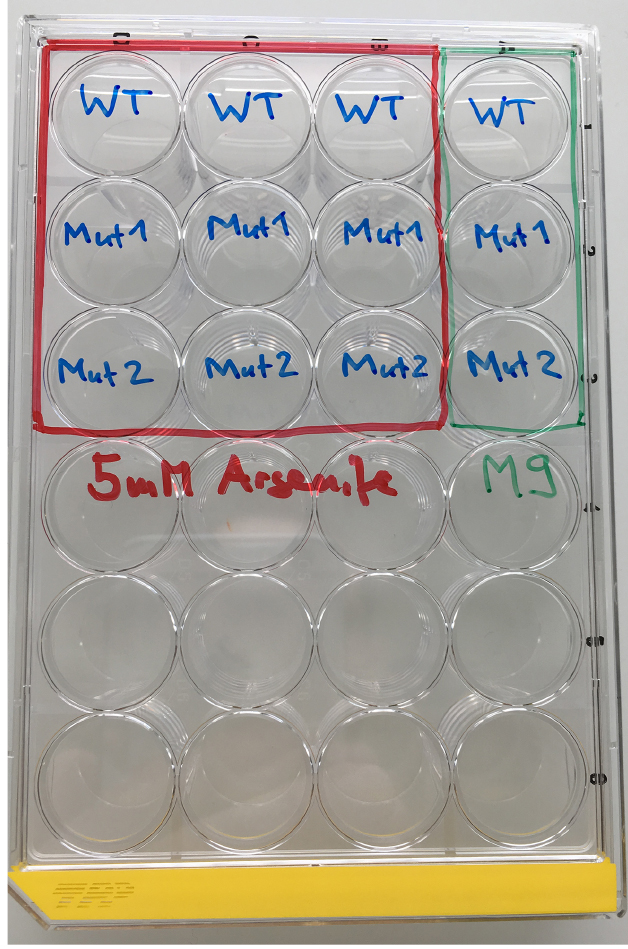
Figure 2. Loading scheme for the As response assay. WT = wild type, Mut = mutant.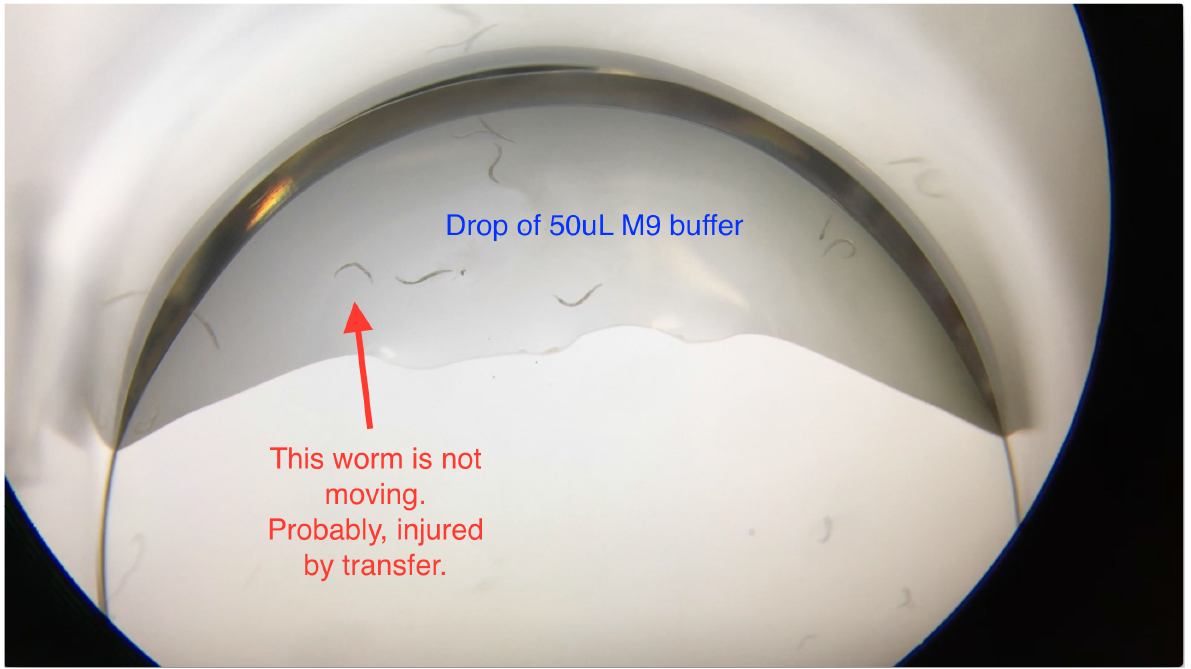
Figure 3. Transferring worms into the drop of M9 buffer in the 24-well plates
Note: When transferring worms into the 50 μl M9 buffer drop in the well, worms may become injured by scratching the worms off the worm pick. Check and exclude non-moving worms (Video 1) before filling up wells with the As solution.
Video 1. Loading C. elegans into 24-well plates for the arsenite oxidative stress assay. 10-12 C. elegans were transferred into a well of the 24-well plate that contains a 50 μl drop of M9. Worms should be freely trashing. Exclude non-moving worms (marked in Figure 3) before filling up wells with the As solution.
Data analysis
For As-assay, the estimates of the survival functions are calculated by using the product-limit (Kaplan-Meier) method (Figure 4 and Table 1). The log-rank (Mantel-Cox) method is used to test the null hypothesis and calculate P-values. Data were analyzed using JMP statistical software from SAS.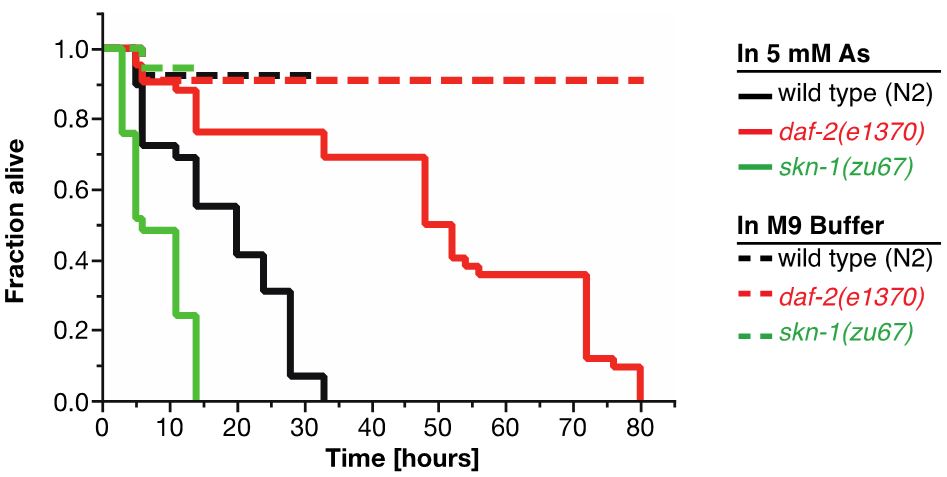
Figure 4. Survival plot of As-assay. Loss-of-function mutation in skn-1 (green curve) makes these animals more sensitive to 5 mM As, whereas reduction-of-function mutation in daf-2 (red curve) makes animals more resistant to 5 mM As (Ewald et al., 2015). For statistical details, please see Table 1.
Table 1. Statistics for As-assay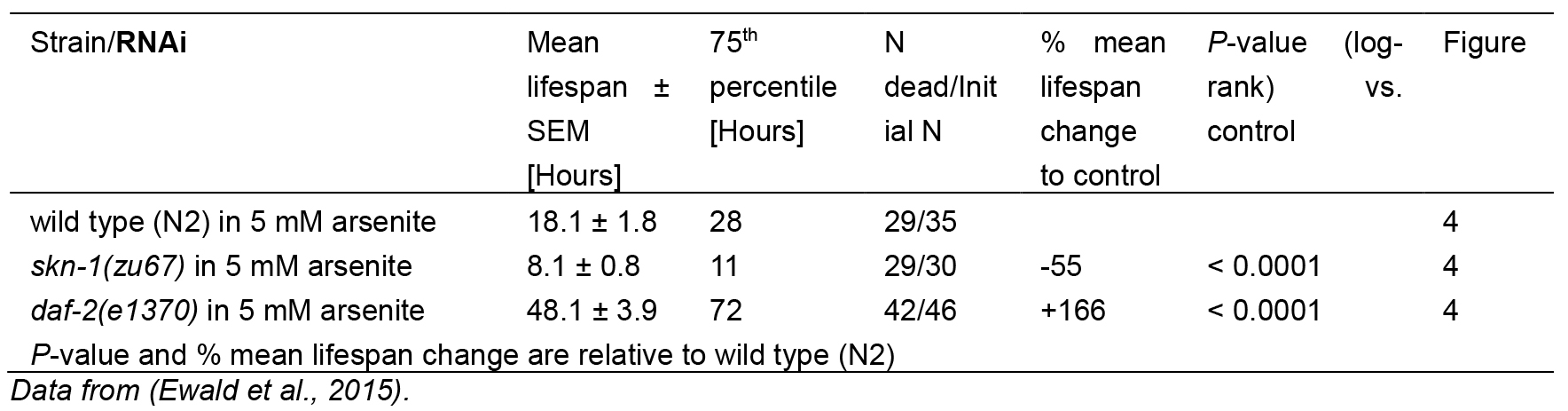
Part II. Protocol for tBHP oxidative stress assay
Materials and Reagents
- Pipette tips
- 6 cm Petri dishes (He, 2011)
- Protective gloves
- Luperox® TBH70X, tert-Butyl hydroperoxide solution 70% in H2O (Sigma-Aldrich, catalog number: 458139 )
Note: tert-Butyl hydroperoxide (tBHP) is an organic peroxide widely used in a variety of oxidation processes. tBHP has an advantage over hydrogen peroxide in that it is less labile. However, tBHP plates should be stored in an airtight container and used within 24 h. tBHP is normally supplied as a 69-70% aqueous solution.
Safety Warning: tert-Butyl hydroperoxide is an exceptionally dangerous chemical that is highly reactive, flammable and toxic. It is corrosive to skin and mucous membranes and causes respiratory distress when inhaled. - Nematode growth medium (NGM) culturing C. elegans plates (He, 2011) seeded with bacteria (OP50) grown overnight (300 μl OP50 per plate)
- Agar (He, 2011)
- Phosphate buffer (He, 2011)
- Calcium chloride (CaCl2) (He, 2011)
- Magnesium sulfate (MgSO4) (He, 2011)
- Cholesterol (He, 2011)
Equipment
- Pipettes
- Face mask
- Safety goggles
- Worm pick
- Fume hood
- Stereomicroscope
Software
- SAS JMP statistical software
Procedure
- Day 0 (Monday): Pick L4 worms onto fresh NGM culturing plate (He, 2011) seeded with OP50 bacteria.
Note: You need 60 L4 worms per strain; 20 worms per tBHP plate, triplicates. - Day 1 (Tuesday): Prepare tBHP plates (see Recipe 1).
- Place agar in dH2O to get a 4% agar solution (100 ml agar = 6 plates) and heat up to solve. Let agar solution cool down (50-60 °C) until you can hold it with your hand and hold it against your wrist, put gloves on, then add all solutions (phosphate buffer, CaCl2, MgSO4, and cholesterol). Shake this briefly to avoid the formation of CaSO4 precipitate and in the end the tBHP in the fume hood. Pour the plates by hand (equal amounts, so that bottom is filled). Place the plates in an airtight box to seal them, and they will be ready for use the next day.
- Day 2 (Wednesday): Pick 20 two-day-old adults onto the tBHP plates (Figure 5). Remember to wear protective gloves, safety goggles, and face mask.

Figure 5. Materials for the tBHP-survival assay - Important: Worms will try to run off the plate (Figure 6). Hence, for the first two hours you need to shuffle worms back into the center of the plate. Therefore, immediately after you put the worms on the tBHP plates, start looking at your first plates again for ‘escaping worms’. We typically assay 4 strains in parallel allowing enough time to shuffle worms back into the center of all 12 plates in a reasonable time.
- Score survival every hour. (Exclude exploded animals from the statistics.)
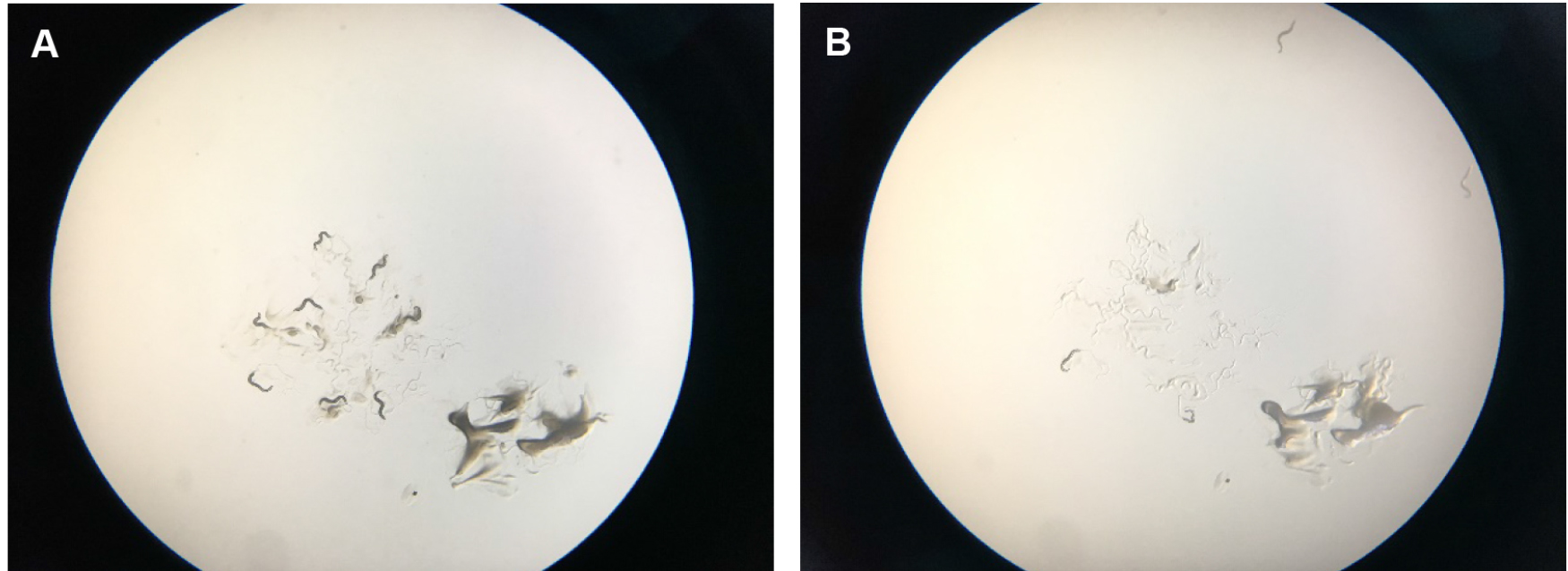
Figure 6. Scoring tBHP plates. A. Shows starting place of worms with a scoop of OP50 bacterial food that attracts them so that they stay a little bit longer in the center of the plate. B. Shows how worms start to run off the plates after several minutes. They try to get away from the tBHP in the plate and usually run off the agar onto the plastic of the petri dish, where they dry out. After two hours, wild-type worms on tBHP will cease crawling around.
Data analysis
For tBHP assay, the estimates of survival functions are calculated by using the product-limit (Kaplan-Meier) method (Figure 7 and Table 2). The log-rank (Mantel-Cox) method is used to test the null hypothesis and calculate P-values. Data were analyzed using JMP statistical software from SAS.
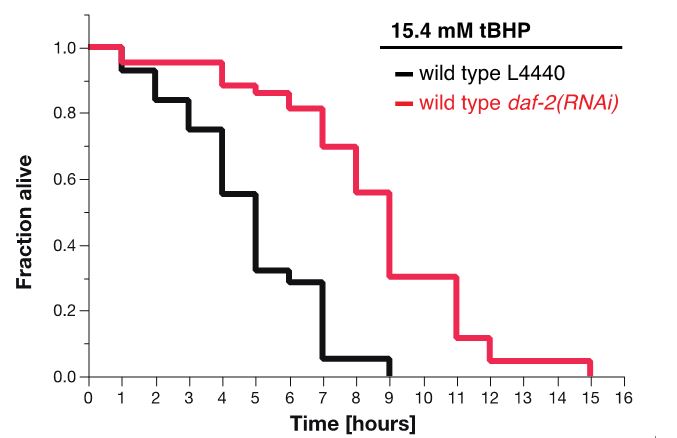
Figure 7. Survival plot of tBHP-assay. RNAi knockdown of daf-2 (red curve) makes wild-type animals more resistant to 15.4 mM tBHP compared to wild-type animals treated with L4440 empty vector control RNAi (Ewald et al., 2015).
Table 2. Statistics for tBHP-assay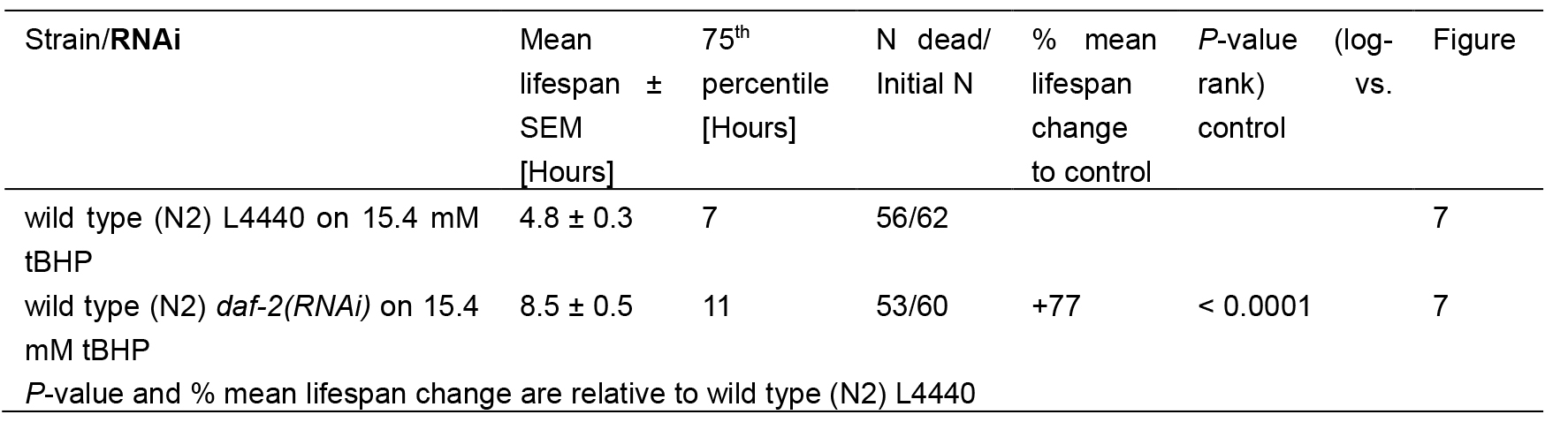
Notes
We want to note that while the protocols described here work reproducibly well, many variations have previously been described, including whether the animals have been provided with food. Different variations can be found in papers from (An and Blackwell, 2003; Tullet et al., 2008; Oliveira et al., 2009; Wang et al., 2010; Robida-Stubbs et al., 2012). The protocols described here are based upon and optimized from this previous work, and have recently been used in (Ewald et al., 2015; Steinbaugh et al., 2015; Hourihan et al., 2016; Ewald et al., 2017).
Recipes
- tBHP plates
100 ml 4% agar dissolved in dH2O
2.5 ml phosphate buffer
100 μl CaCl2
100 μl MgSO4
160 μl cholesterol
214 μl tBHP [Stock = 70%] hence, final concentration = 15.4 mM
Acknowledgments
We thank the Blackwell and Ewald lab for developing and refining these assays. Picture and movie credit for Nadine Herrmann and Eline Jongsma (ETH Zurich).
References
- An, J. H. and Blackwell, T. K. (2003). SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17(15): 1882-1893.
- Blackwell, T. K., Steinbaugh, M. J., Hourihan, J. M., Ewald, C. Y. and Isik, M. (2015). SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med 88(Pt B): 290-301.
- Ewald, C. Y., Hourihan, J. M., Bland, M. S., Obieglo, C., Katic, I., Moronetti Mazzeo, L. E., Alcedo, J., Blackwell, T. K. and Hynes, N. E. (2017). NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans. Elife 6.
- Ewald, C. Y., Landis, J. N., Porter Abate, J., Murphy, C. T. and Blackwell, T. K. (2015). Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 519(7541): 97-101.
- He, F. L. (2011). Common worm media and buffers. Bio-protocol Bio101: e55.
- Hourihan, J. M., Moronetti Mazzeo, L. E., Fernandez-Cardenas, L. P. and Blackwell, T. K. (2016). Cysteine sulfenylation directs IRE-1 to activate the SKN-1/Nrf2 antioxidant response. Mol Cell 63(4): 553-566.
- Oliveira, R. P., Porter Abate, J., Dilks, K., Landis, J., Ashraf, J., Murphy, C. T. and Blackwell, T. K. (2009). Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8(5): 524-541.
- Robida-Stubbs, S., Glover-Cutter, K., Lamming, D. W., Mizunuma, M., Narasimhan, S. D., Neumann-Haefelin, E., Sabatini, D. M. and Blackwell, T. K. (2012). TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab 15(5): 713-724
- Steinbaugh, M. J., Narasimhan, S. D., Robida-Stubbs, S., Moronetti Mazzeo, L. E., Dreyfuss, J. M., Hourihan, J. M., Raghavan, P., Operana, T. N., Esmaillie, R. and Blackwell, T. K. (2015). Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. Elife 4.
- Tullet, J. M., Hertweck, M., An, J. H., Baker, J., Hwang, J. Y., Liu, S., Oliveira, R. P., Baumeister, R. and Blackwell, T. K. (2008). Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132(6): 1025-1038.
- Wang, J., Robida-Stubbs, S., Tullet, J. M., Rual, J. F., Vidal, M. and Blackwell, T. K. (2010). RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet 6(8).
Article Information
Copyright
Ewald et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ewald, C. Y., Hourihan, J. M. and Blackwell, T. K. (2017). Oxidative Stress Assays (arsenite and tBHP) in Caenorhabditis elegans. Bio-protocol 7(13): e2365. DOI: 10.21769/BioProtoc.2365.
- Ewald, C. Y., Hourihan, J. M., Bland, M. S., Obieglo, C., Katic, I., Moronetti Mazzeo, L. E., Alcedo, J., Blackwell, T. K. and Hynes, N. E. (2017). NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans. Elife 6.
Category
Developmental Biology > Cell signaling > Stress response
Neuroscience > Behavioral neuroscience > Animal model > Other
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










