- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Active Cdk5 Immunoprecipitation and Kinase Assay
Published: Vol 7, Iss 13, Jul 5, 2017 DOI: 10.21769/BioProtoc.2363 Views: 10095
Reviewed by: Pengpeng LiLiang LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

ZnCl2 Precipitation-Assisted Sample Preparation for Proteomic Analysis
Qiqing He [...] Fuchu He
Jul 20, 2025 2733 Views

Protocol for the Preparation of a Recombinant Treacle Fragment for Liquid–Liquid Phase Separation (LLPS) Assays
Nadezhda V. Petrova [...] Artem K. Velichko
Sep 20, 2025 1823 Views

Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
Basil Baby Mattamana [...] Peter Allen Faull
Dec 20, 2025 1261 Views
Abstract
Cdk5 activity is regulated by the amounts of two activator proteins, p35 and p39 (Tsai et al., 1994; Zheng et al., 1998; Humbert et al., 2000). The p35-Cdk5 and p39-Cdk5 complexes have differing sensitivity to salt and detergent concentrations (Hisanaga and Saito, 2003; Sato et al., 2007; Yamada et al., 2007; Asada et al., 2008). Cdk5 activation can be directly measured by immunoprecipitation of Cdk5 with its bound activator, followed by a Cdk5 kinase assay. In this protocol, buffers for cell lysis and immunoprecipitation are intended to preserve both p35- and p39-Cdk5 complexes to assess total Cdk5 activity. Cells are lysed and protein concentration is determined in the post-nuclear supernatant. Cdk5 is immunoprecipitated from equal amounts of total protein between experimental groups. Washes are then performed to remove extraneous proteins and equilibrate the Cdk5-activator complexes in the kinase buffer. Cdk5 is then incubated with histone H1, a well-established in vitro target of Cdk5, and [γ-32P]ATP. Reactions are resolved by SDS-PAGE and transferred to membranes for visualization of H1 phosphorylation and immunoblot of immunoprecipitated Cdk5 levels. We have used this assay to establish p39 as the primary activator for Cdk5 in the oligodendroglial lineage. However, this assay is amenable to other cell lineages or tissues with appropriate adjustments made to lysis conditions.
Keywords: Kinase assayBackground
Although Cdk5 is typically associated with neuronal function, recent work has demonstrated that Cdk5 also regulates oligodendroglia progenitor cell (OPC) development (Tang et al., 1998; Miyamoto et al., 2007 and 2008). Cdk5 function is critical for OPC migration and differentiation, and loss of Cdk5 results in CNS hypomyelination (Miyamoto et al., 2007 and 2008; He et al., 2010; Yang et al., 2013). However, molecular mechanisms that regulate Cdk5 function in neurons and OLs remain elusive. The activity of Cdk5 is controlled by the available amounts of two activator homologs, p35 and p39 (Tsai et al., 1994; Zheng et al., 1998; Humbert et al., 2000). The defects in embryonic brain development and perinatal lethality observed in mice lacking both p35 and p39 were nearly identical to defects in the Cdk5-null mice (Ohshima et al., 1996; Ko et al., 2001), indicating that p35 and p39 are the sole activators of Cdk5 in the brain. We uncovered that in contrast to the major role of p35 in activating Cdk5 in neurons, p39 is the primary Cdk5 activator in oligodendrocytes (OLs), where p35 expression is negligible. Using this active Cdk5 immunoprecipitation and kinase assay, we demonstrated that Cdk5 activity is almost completely ablated in OLs with siRNA-mediated p39 knockdown. Previous work established the differing sensitivity of p35 and p39 to high detergent and salt concentrations (Hisanaga and Saito, 2003; Sato et al., 2007; Yamada et al., 2007; Asada et al., 2008). Based on those reports, this protocol was developed to try and preserve both p35- and p39-Cdk5 complexes to measure total Cdk5 activity regardless of activator. Our work further showed that p39 is essential for OL differentiation and myelin repair, with upregulation of p35 masking the loss of p39 function during myelin development. Measuring Cdk5 activity from cells, in combination with immunoblots for Cdk5 target phosphorylation, provides a tool to identify novel regulators of Cdk5 activation.
Materials and Reagents
- Cell lifter (Corning, catalog number: 3008 )
- Corning sterile 60 mm cell culture dishes (Corning, catalog number: 3261 )
- Corning sterile 100 mm cell culture dishes (Corning, catalog number: 3262 )
- 15 ml conical tubes (Denville Scientific, catalog number: C1018-P )
- Micro slides (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4951PLUS4 )
- Micro cover glass (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 25X54I24901 )
- 1.7 ml tubes (Denville Scientific, SlipTechTM, catalog number: C19033 )
- 50 ml conical tubes (Denville Scientific, catalog number: 1005513 )
- Nitrocellulose or PVDF membrane (GE Healthcare, catalog number: RPN82D or 10600021 )
- Autoclaved micropipette tips (Denville Scientific, Woodpecker ReloadsTM, catalog numbers: P2102-NB , P2101-N , P2109 )
- X-ray film (Denville Scientific, Hyblot ES®, catalog number: E3218 )
- Trypan blue (Sigma-Aldrich, catalog number: 302643 )
- BCA Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 23235 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A2153 )
- Antibody against Cdk5 (Santa Cruz C-8) (Santa Cruz Biotechnology, catalog number: sc-173 )
- Antibody against Cdk5 (Cell Signaling Technology, catalog number: 2506 )
- Rabbit IgG antibody (Vector Laboratories, catalog number: I-1000 )
- Goat anti-rabbit-horseradish peroxidase (HRP) (Jackson ImmunoResearch, catalog number: 111-035-003 )
- 10 mM ATP (Thermo Fisher Scientific, catalog number: PV3227 )
- Cdk5/p25, active complex (EMD Millipore, catalog number: 14-516 )
- [γ-32P]ATP (PerkinElmer, catalog number: BLU002Z250UC )
- Histone H1 (EMD Millipore, catalog number: 14-155 )
- 12% polyacrylamide gel (Bio-Rad Laboratories, catalog number: 4561043 )
- Enhanced Chemiluminescence Reagent Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 32106 )
- Tris/Glycine/SDS buffer (Bio-Rad Laboratories, catalog number: 1610732 )
- Methanol (Sigma-Aldrich, catalog number: 34860 )
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S7907 )
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Tris base (Sigma-Aldrich, catalog number: RDD008 )
- Concentrated HCl (Sigma-Aldrich, catalog number: 295426 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: EDS )
- Sodium hydroxide (NaOH) tablets (Sigma-Aldrich, catalog number: S8045 )
- Ethylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889 )
- 3-(N-morpholino)propanesulfonic acid (MOPS) (Sigma-Aldrich, catalog number: RDD003 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- Sodium fluoride (NaF) (Sigma-Aldrich, catalog number: S7920 )
- NP-40/IGEPAL® CA-630 (Sigma-Aldrich, catalog number: I3021 )
- Polymethyl sulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626 )
- 100% ethanol (Sigma-Aldrich, catalog number: E7023 )
- Pepstatin (Sigma-Aldrich, catalog number: P4265 )
- Leupeptin (Sigma-Aldrich, catalog number: L2884 )
- Aprotinin (Sigma-Aldrich, catalog number: A1153 )
- Protein A Sepharose CL-4B (GE Healthcare, catalog number: 17-0780-01 )
- Pre-activated sodium orthovanadate (100 mM Na3VO4) (New England Biolabs, catalog number: P0758L )
- 10% BrijTM-35 (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 28316 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- 2-mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- Magnesium acetate (MgOAc) (Sigma-Aldrich, catalog number: M5661 )
- Tween-20 (Sigma-Aldrich, catalog number: P1379 )
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126 )
- 1x transfer buffer (see Recipes)
- 10x phosphate-buffered saline (PBS) (see Recipes)
- 1x phosphate-buffered saline (PBS) (see Recipes)
- 2 M Tris-HCl (pH 7.5) (see Recipes)
- 50 mM Tris-HCl (pH 7.5) (see Recipes)
- 5 M NaCl (see Recipes)
- 0.5 M EDTA (pH 8.0) (see Recipes)
- 0.5 M EGTA (pH 8.0) (see Recipes)
- 1 M MOPS (pH 7.0) (see Recipes)
- 1 M MgCl2 (see Recipes)
- 1 M NaF (see Recipes)
- 20% NP-40 (see Recipes)
- 100 mM PMSF (see Recipes)
- 1 mg/ml pepstatin A (see Recipes)
- 1 mg/ml leupeptin (see Recipes)
- 1 mg/ml aprotinin (see Recipes)
- 50% slurry of Protein A Sepharose CL-4B (see Recipes)
- Cdk5 lysis buffer (stock) (see Recipes)
- Cdk5 lysis buffer (working) (see Recipes)
- Cdk5 kinase buffer (stock) (see Recipes)
- Cdk5 kinase buffer (washes) (see Recipes)
- Cdk5 kinase buffer (assay) (see Recipes)
- MOPS dilution buffer (see Recipes)
- 5x reaction buffer (see Recipes)
- 50 mM magnesium acetate buffer (MgOAc) (see Recipes)
- 1x phosphate-buffered saline/0.1% Tween-20 (PBS-T) (see Recipes)
- 5x Laemmli buffer (see Recipes)
- 0.2% bromophenol blue (see Recipes)
Equipment
- Hemacytometer (Hausser Scientific, catalog number: 3100 )
- PIPETMAN ClassicTM pipets (Gilson, model: P10, catalog number: F144802 )
- PIPETMAN ClassicTM pipets (Gilson, model: P20, catalog number: F123600 )
- PIPETMAN ClassicTM pipets (Gilson, model: P200, catalog number: F123601 )
- PIPETMAN ClassicTM pipets (Gilson, model: P1000, catalog number: F123602 )
- Refrigerated tabletop centrifuge for 15 ml conical tubes (Jouan, model: CT422 )
- Tabletop centrifuge for 1.5 ml tubes in a 4 °C cold room (Eppendorf, model: 5415 D )
- Inverted light microscope (Olympus, model: CK30 )
- Certified Geiger counter (W.B. Johnson Instruments, model: GSM-110 )
- Plexiglass shielding (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 6700-1812 )
- Phosphorimaging cassette (Thomas Scientific, catalog number: C993J84)
Manufacture: bioWORLD, catalog number: 43121008-1 . - Autoradiography cassette (Denville Scientific, catalog number: E3122 )
- 500 ml glass bottles (Corning, PYREX®, catalog number: 1397-500 )
Software
- GraphPad Prism (GraphPad Software)
- ImageJ (https://imagej.nih.gov/ij)
Procedure
Notes:
- Steps A1-A4 should be performed as quickly as possible to avoid loss of cellular proteins during washes.
- This protocol is designed to preserve the Cdk5/activator complex while still maintaining enough stringency to reasonably lyse the cells and remove non-specific proteins from the immunoprecipitated complex.
- Once [γ-32P]ATP has been added in step D3b, ALL discarded solutions MUST be disposed of according to the radiation safety protocols at your institution for the remainder of this protocol.
- Generating cellular lysates
- Cell cultures should be grown to confluence with the appropriate treatment and control groups.
- Aspirate the media from the plate(s) and rinse the cell cultures very gently with PBS (see Recipes) twice. Remove the PBS from the last wash.
- Detach the cells from the culture plates using a cell lifter. Collect the detached cells by rinsing the plate with an appropriate volume of PBS (1 ml for 60 mm plates, 3 ml for 100 mm plates, etc.).
- Transfer the cell suspension to a 15 ml conical tube.
- Centrifuge the cell suspension at 3,500 x g for 5 min at 4 °C.
- Repeat steps A3-A5 if needed.
- Resuspend the cell pellets in lysis buffer (see Recipes) at 15% w/v.
- Incubate lysates on ice for 10 min with periodic agitation.
- Confirm lysis by mixing a 10 µl sample of the lysate 1:1 with trypan blue. Pipette 10 µl of this mixture onto a microscope slide and place a coverslip onto the 10 µl droplet. Good lysis will appear as many intact nuclei that are no longer contained within cells. Unlysed cells will appear as spheres with blue outlines. If the nuclei lyse, many brown fibers will be present, which can negatively impact the purity of your immunoprecipitation. Example images of unlysed cells and good lysis are presented in Figure 1.
- Reserve 10% of the lysate as ‘Input’, add 5x Laemmli buffer (see Recipes) to Input to a final concentration of 1x, and store samples on ice until step D9.
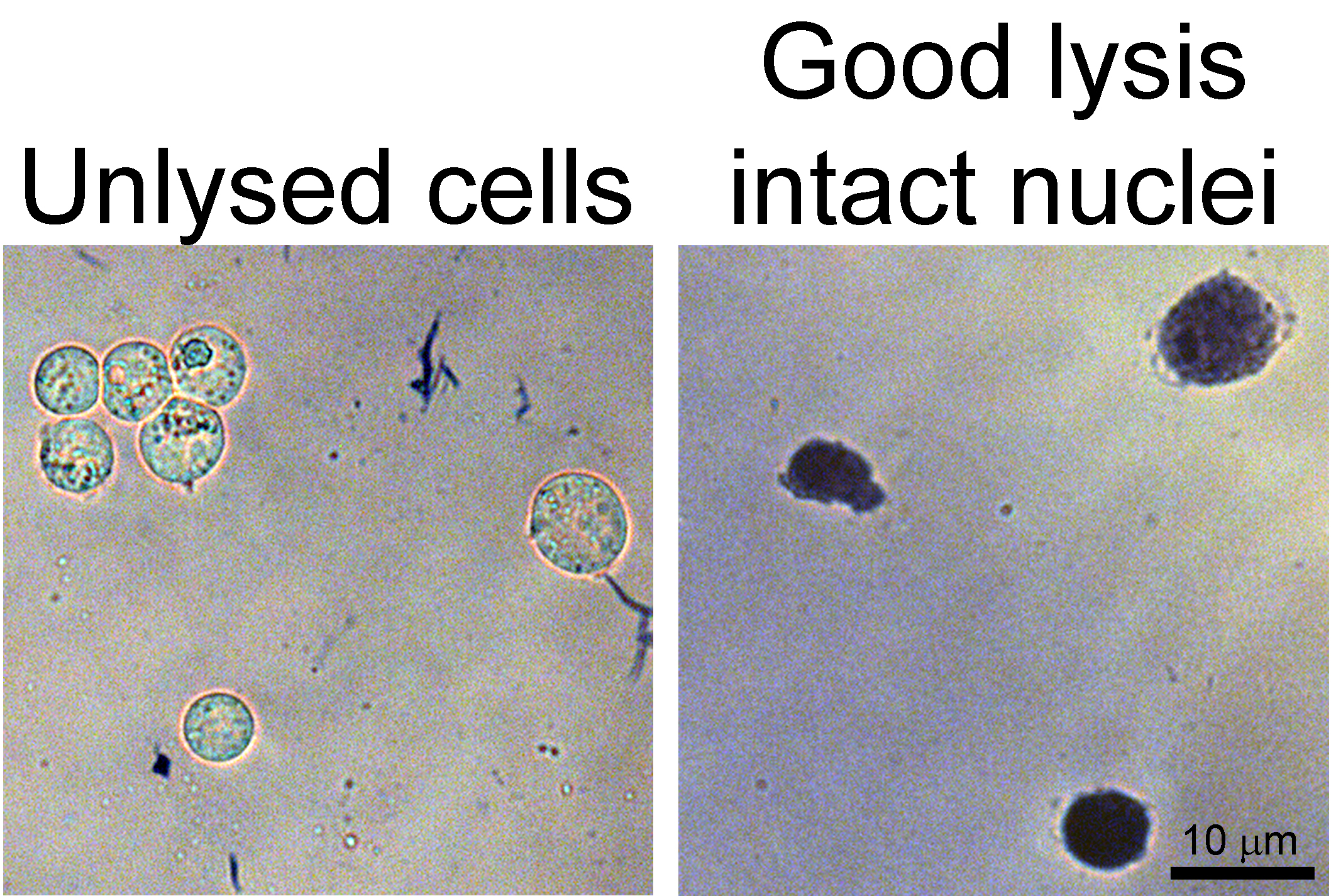
Figure 1. Lysis confirmation. Left, example image of unlysed cells stained with trypan blue. Note the blue outline and slight halo around each cell. Right, example image of intact nuclei from lysed cells.
- Cell cultures should be grown to confluence with the appropriate treatment and control groups.
- Preparing sample for immunoprecipitation
- Centrifuge the lysate at 10,000 x g for 10 min at 4 °C.
- Transfer the supernatant to a fresh tube.
- Add 50 µl of 50% slurry of Protein A beads (or whichever type of beads you will use for the immunoprecipitation) to the lysate.
- Rotate or rock the mixture of beads and lysate at 4 °C for 30 min to pre-clear the lysate of proteins that non-specifically bind to the beads.
- Centrifuge the mixture at 3,000 x g for 2 min at 4 °C.
- Transfer the supernatant to a fresh tube.
- Quantify the concentration of protein in the lysate using standard protein quantification methods (Bradford or BCA kits, for example)
- Generate a standard curve of 1 mg/ml, 0.5 mg/ml, 0.25 mg/ml, 0.125 mg/ml, and 0 mg/ml bovine serum albumin (BSA) by diluting it using sterile water.
- Dilute the lysate (between 1:2 to 1:10 for cell lysate) so that the sample protein concentration will fall within the range of the BSA standard curve.
- Follow the manufacturer’s instructions to record the concentration of protein in the lysate. Take note of the R2 value as it indicates the accuracy of the protein concentration quantification (0.9 and above is desired).
- Take the average of three readings.
- Centrifuge the lysate at 10,000 x g for 10 min at 4 °C.
- Immunoprecipitating the Cdk5/activator complex
- Prepare two fresh 1.5 ml tubes with 1 mg of total protein from the lysate.
Note: If one of your samples contains insufficient protein concentration, equalize the total protein amount in all samples to match the protein amounts possible from the sample with the lowest protein concentration. - Add lysis buffer to bring the total volume in each tube to 500 µl.
- Add 10 µl (2 µg) Santa Cruz anti-Cdk5 antibody (C-8) to one of the 1.5 ml tubes prepared for each of your samples.
- Add 2 µg of IgG antibody to the other 1.5 ml tubes prepared for each of your samples.
- Rotate or rock the mixture of beads and lysate at 4 °C for 3 h.
- Add 50 µl of 50% slurry of Protein A beads to each tube.
- Rotate or rock the mixture of beads and lysate at 4 °C for 2 h.
- Centrifuge the tubes at 3,000 x g for 2 min at 4 °C.
- Remove and discard the supernatant.
- Add 500 µl of lysis buffer containing 0.1% NP-40.
- Rotate or rock the mixture of beads and lysate at 4 °C for 10 min.
- Repeat steps C8-C11 twice more.
- Centrifuge the tubes at 3,000 x g for 2 min at 4 °C.
- Remove and discard the supernatant.
- Add 500 µl of kinase buffer (see Recipes) without ATP or histone H1.
- Rotate or rock the mixture of beads and lysate at 4 °C for 10 min.
- Repeat steps C13-C16 once more (general scheme of immunoprecipitation in Figure 2).
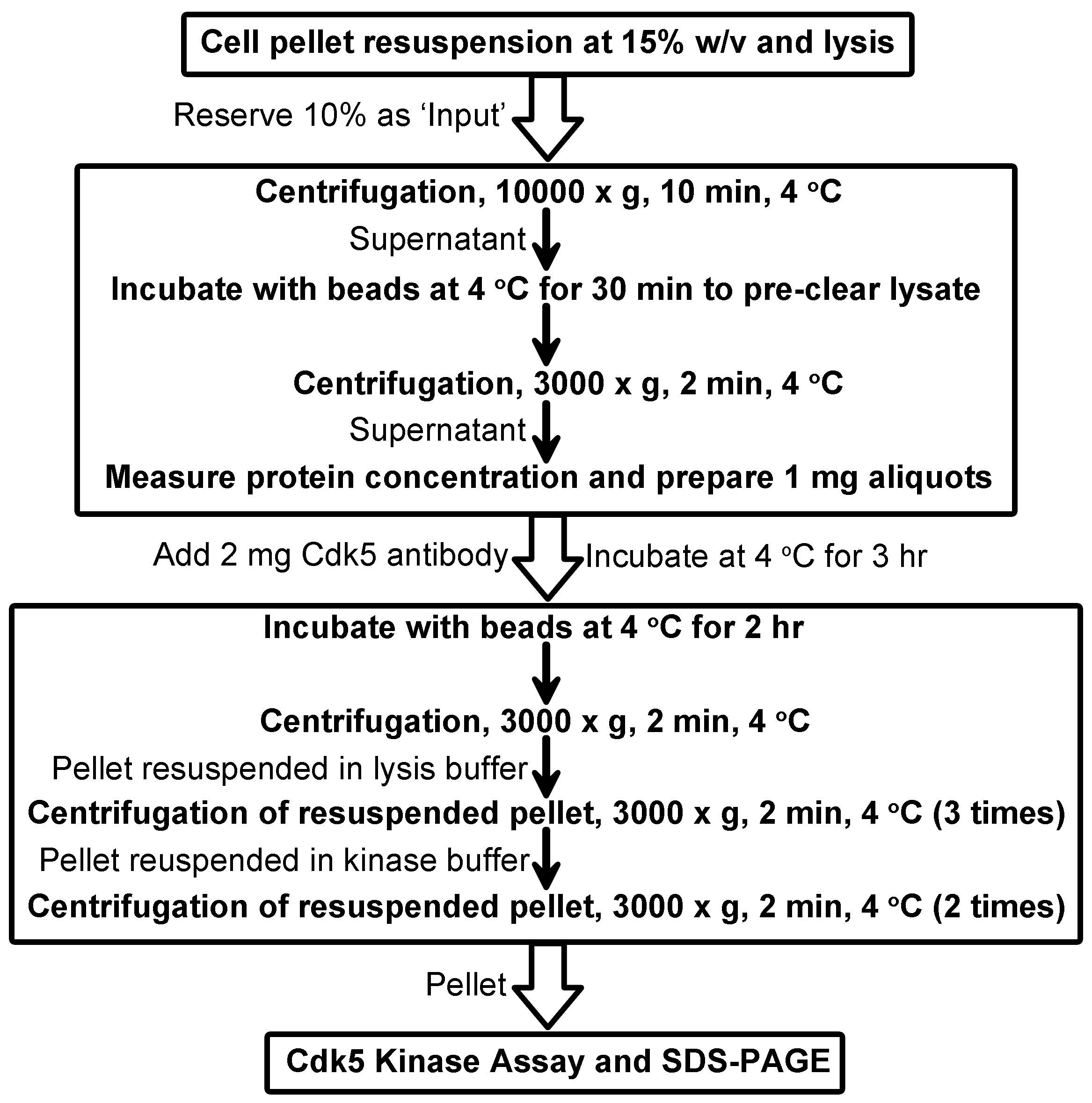
Figure 2. Scheme for immunoprecipitation of active Cdk5 complexes
- Prepare two fresh 1.5 ml tubes with 1 mg of total protein from the lysate.
- Cdk5 kinase assay
- Resuspend beads in 25 µl kinase buffer (with 100 ng/µl H1 and 25 µM ATP).
- Prepare a reaction with recombinant Cdk5/p25 for a positive control (general scheme for Cdk5 kinase assay and controls in Figure 3).
- Add 14.6 µl MOPS dilution buffer (see Recipes) to 1 µl Cdk5/p25 activated complex (Upstate/Millipore).
- Add 5 µl 5x reaction buffer (see Recipes) to a fresh 1.5 ml tube.
- Add 2.5 µl of histone H1 (final 100 ng/µl).
- Add 2.5 µl of diluted Cdk5 from step D2a to the 1.5 ml tube.
- Add 5 µl of sterile water.
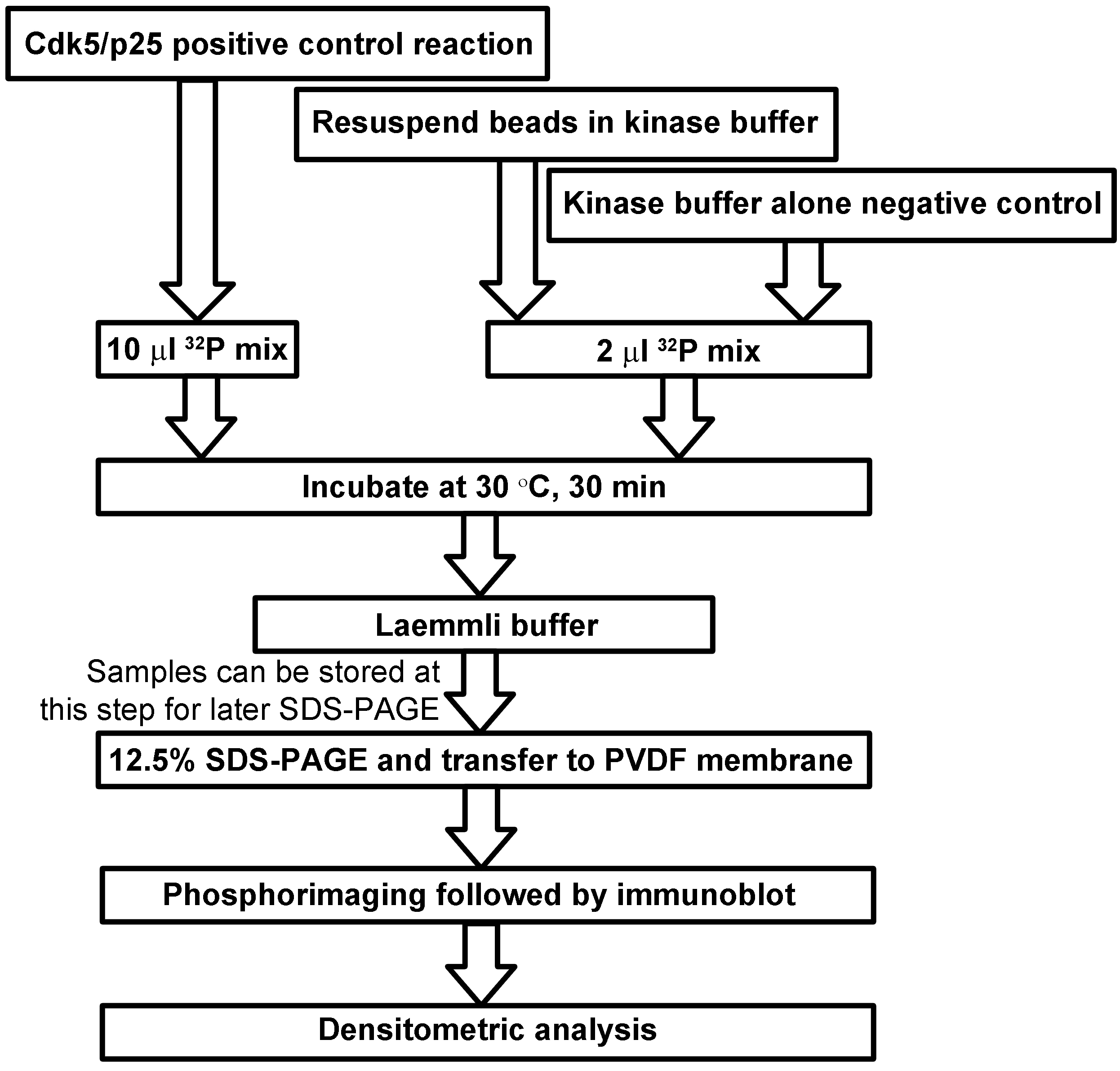
Figure 3. Scheme for Cdk5 kinase assay, SDS-PAGE, and detection
- Add 14.6 µl MOPS dilution buffer (see Recipes) to 1 µl Cdk5/p25 activated complex (Upstate/Millipore).
- Prepare a working solution of [γ-32P]ATP
- Prepare 47 µl of 25 mM magnesium acetate (see Recipes).
- Add [γ-32P]ATP (final concentration 0.1 µCi/µl).
- Add 10 mM ATP to bring the total volume to 50 µl.
- Prepare 47 µl of 25 mM magnesium acetate (see Recipes).
- Add 10 µl of the 32P mix from step D3 to the positive control reaction prepared in step D2.
- Add 2 µl of 32P mix from step D3 to each IP reaction from step D1.
- Mix 25 µl of kinase buffer with 2 µl of 32P mix from step D3 as a negative control.
- Incubate all reactions at 30 °C for 30 min.
- Add 5 µl of 5x Laemmli buffer to each reaction.
- OPTIONAL: Store reactions at -20 °C.
- Resolve immediately by 12.5% SDS-PAGE.
- DO NOT allow the dye front to exit the bottom of the gel. This will minimize radioactive contamination of the running buffer or equipment.
- After resolving reactions, cut off dye at bottom (contains free 32P) and stacking gel.
- Transfer the resolved proteins to nitrocellulose or PVDF membrane.
- Expose the membrane to film or phosphorscreen at -80 °C (see example in Figure 4).
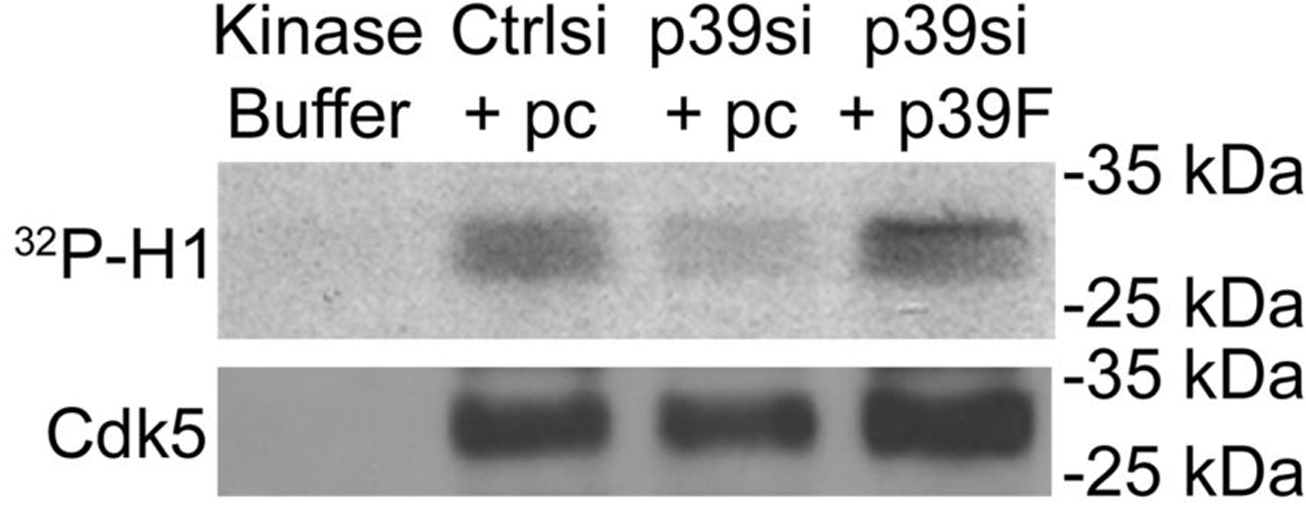
Figure 4. Detection of Cdk5 kinase activity and protein levels in immunoprecipitates. Results of Cdk5 immunoprecipitation and kinase assay performed from cells transfected with control siRNA with empty vector (Ctrlsi + pc), p39 siRNA with empty vector (p39si + pc), p39 siRNA with p39-FLAG (p39si + p39F). Kinase buffer alone was included as a negative control. In this figure, the exogenously expressed p39-FLAG reversed the effect on Cdk5 activity caused by siRNA-mediated knockdown of endogenous p39. This research was originally published in Journal of Biological Chemistry. Bankston AN, Li W, Zhang H, Ku L, Liu G, Papa F, Zhao L, Bibb JA, Cambi F, Tiwari-Woodruff SK, Feng Y. p39, the primary activator for cyclin-dependent kinase 5 (Cdk5) in oligodendroglia, is essential for oligodendroglia differentiation and myelin repair. J Biol Chem. 2013; 288(25):18047-57. © the American Society for Biochemistry and Molecular Biology.
- Resuspend beads in 25 µl kinase buffer (with 100 ng/µl H1 and 25 µM ATP).
- Performing Western blot to assess immunoprecipitation
- Incubate the membrane for one hour with 10% non-fat milk in PBS-T (see Recipes).
- Discard 10% milk solution.
- Incubate the membrane with 2% non-fat milk in PBS-T containing a 1:1,000 dilution of anti-Cdk5 antibody (Cell Signaling Technology) at 4 °C overnight.
- Discard the 2% milk solution containing anti-Cdk5 antibody.
- Wash the membrane by incubating it in PBS-T for 10 min.
- Discard the PBS-T wash solution.
- Repeat steps E5-E6 twice more.
- Incubate the membrane with 2% non-fat milk in PBS-T containing a 1:5,000 dilution of HRP-conjugated anti-rabbit IgG antibody (Jackson ImmunoResearch) at room temperature for one hour.
- Wash the membrane by incubating it in PBS-T for 10 min.
- Discard the PBS-T wash solution.
- Incubate the membrane with an enhanced chemiluminescence solution according to the manufacturer’s protocol.
- Expose the membrane to film (see example in Figure 4).
- Incubate the membrane for one hour with 10% non-fat milk in PBS-T (see Recipes).
Data analysis
- For experimental design, the immunoprecipitation and kinase assay is performed a minimum of three times for a given experiment.
- Densitometric analysis is performed using ImageJ. For each sample the intensity of histone H1 phosphorylation measured in step D13 is normalized to the intensity of Cdk5 protein measured in step E12 (see Figure 5).
- For statistical analysis, we use GraphPad Prism software. To compare two samples, a two-tailed Student’s t-test was used. To compare three or more samples, one-way ANOVA with a post-hoc Tukey correction was used.
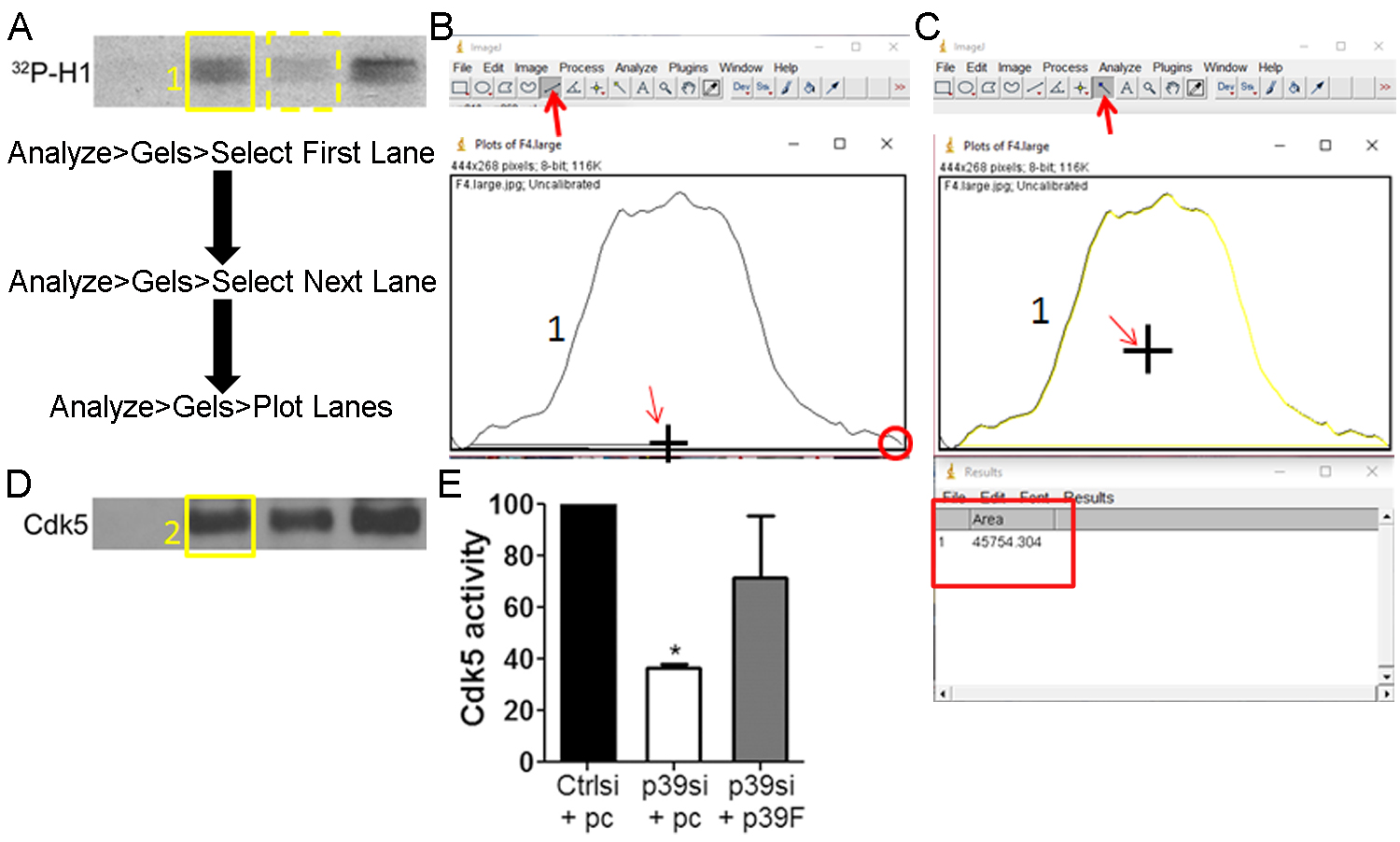
Figure 5. Measurement and normalization of Cdk5 kinase activity and protein levels in ImageJ. A. Draw a box around the first lane of Cdk5 kinase activity (yellow solid box). Click Select First Lane under the Analyze Gels menu. Move the box over the second lane of Cdk5 kinase activity (yellow dashed box). Click Select Second Lane under the Analyze Gels menu. Move the box over each of the remaining lanes of Cdk5 kinase activity, clicking Select Second Lane under the Analyze Gels menu at each lane. Click Plot Lanes under the Analyze Gels menu. B. Plots will be generated for each lane. Use the line tool (indicated by bold red arrow) to draw a line across the bottom of the plot (cursor drawing line indicated by slim red arrow). Be sure to close any gaps (red circle). Do this for all graphs generated. C. Use the magic wand tool (indicated by bold red arrow) to measure the area within the intensity plot by clicking inside the enclosed plot area (cursor indicated by slim red arrow). Do this for all graphs generated. A Results table will be generated with each row corresponding to each lane. D. Repeat the steps in panels A-C for the Cdk5 immunoblot. E. Divide the area result from each lane of Cdk5 kinase activity by the area result for the same lane of the Cdk5 immunoblot. Graph and analyze as needed.
Notes
- Consult your institution’s radiation safety office before beginning this protocol. They can help you set up an isolated station for handling the radioactive materials as well as teach proper technique. They also likely have required protocols for ordering and disposing of radioactive materials.
- The detergent type or concentration in the lysis buffer may have to be modified to efficiently lyse your cells of interest. Time of lysis may also be an important factor. All subsequent buffers should contain 0.1% NP-40 to preserve the Cdk5/activator complexes.
- We include two obligate controls in this protocol: 1) active Cdk5/p25 complex as a positive control; 2) kinase buffer only group as a negative control to ensure reagent purity. Other controls which are specific to your experiment should be considered.
- This assay should be used in combination with Western blot for phosphorylated and total levels of known Cdk5 targets in lysates of your cells to confirm any changes observed in Cdk5 activity.
- When rescuing expression of a target of siRNA-mediated knockdown (for example, p39 in this protocol), it is optimal to use an expression vector (p39F) that contains silent point mutations in the sequence which is targeted by the siRNA (p39si). This will make the rescue expression vector resistant to the siRNA.
Recipes
- 1x transfer buffer (1,000 ml)
- Weigh 3.03 g of Tris-HCl
- Add 14.4 g of glycine
- Add 700 ml of sterile water
- Add 200 ml of methanol
- Add sterile water to a final volume of 1,000 ml
- Weigh 3.03 g of Tris-HCl
- 10x phosphate-buffered saline (PBS) (1,000 ml)
- Weigh 14.4 g of Na2HPO4, 2.4 g KH2PO4, 2.0 g KCl, and 80.0 g NaCl
- Add 800 ml of sterile water
- Stir until dissolved
- Bring the final volume to 1,000 ml using sterile water
- Weigh 14.4 g of Na2HPO4, 2.4 g KH2PO4, 2.0 g KCl, and 80.0 g NaCl
- 1x phosphate-buffered saline (PBS) (500 ml)
- Add 50 ml of 10x PBS
- Bring final volume to 500 ml using sterile water
- Add 50 ml of 10x PBS
- 2 M Tris-HCl (pH 7.5) (500 ml)
- Weigh 121.14 g of Tris-base powder in a 500 ml glass bottle
- Add 450 ml of sterile water
- Stir until dissolved
- Adjust the pH to 7.5 using concentrated HCl
- Bring the final volume to 500 ml using sterile water
- Weigh 121.14 g of Tris-base powder in a 500 ml glass bottle
- 50 mM Tris-HCl (pH 7.5) (50 ml)
- Add 1.25 ml of 2 M Tris-HCl (pH 7.5)
- Bring final volume to 50 ml using sterile water
- Add 1.25 ml of 2 M Tris-HCl (pH 7.5)
- 5 M NaCl (500 ml)
- Weigh 146.1 g of NaCl in a 500 ml glass bottle
- Add 450 ml of sterile water
- Stir until dissolved
- Bring the final volume to 500 ml using sterile water
- Weigh 146.1 g of NaCl in a 500 ml glass bottle
- 0.5 M EDTA (pH 8.0) (50 ml)
- Weigh 7.31 g of EDTA powder in a 50 ml conical tube
- Add 450 ml of sterile water
- Stir and heat until dissolved
- Adjust the pH to 8.0 using NaOH tablets to aid dissolution
- Bring the final volume to 50 ml using sterile water
- Weigh 7.31 g of EDTA powder in a 50 ml conical tube
- 0.5 M EGTA (pH 8.0) (50 ml)
- Weigh 9.51 g of EGTA powder in a 50 ml conical tube
- Add 450 ml of sterile water
- Stir and heat until dissolved
- Adjust the pH to 8.0 using NaOH tablets to aid dissolution
- Bring the final volume to 50 ml using sterile water
- Weigh 9.51 g of EGTA powder in a 50 ml conical tube
- 1 M MOPS (pH 7.0) (500 ml)
- Weigh 104.63 g of MOPS in a 500 ml glass bottle
- Add 450 ml of sterile water
- Stir until dissolved
- Adjust pH to 7.0 using NaOH tablets
- Bring the final volume to 500 ml using sterile water
- Weigh 104.63 g of MOPS in a 500 ml glass bottle
- 1 M MgCl2 (10 ml)
- Weigh 0.95 g of MgCl2 in a 15 ml conical tube
- Add 9.5 ml of sterile water
- Shake until dissolved
- Bring the final volume to 10 ml using sterile water
- Weigh 0.95 g of MgCl2 in a 15 ml conical tube
- 1 M NaF (10 ml)
- Weigh 0.42 g of NaF in a 15 ml conical tube
- Add 9.5 ml of sterile water
- Shake until dissolved
- Bring the final volume to 10 ml using sterile water
- Weigh 0.42 g of NaF in a 15 ml conical tube
- 20% NP-40
- Mix 2 ml NP-40 with 8 ml sterile water
- Vortex and heat at 30 °C until well mixed
- Mix 2 ml NP-40 with 8 ml sterile water
- 100 mM PMSF (1 ml)
- Weigh 0.017 g of PMSF in a 1.5 ml tube
- Add 1 ml 100% ethanol
- Mix until dissolved
- Weigh 0.017 g of PMSF in a 1.5 ml tube
- 1 mg/ml pepstatin A (1 ml)
- Weigh 1 mg of pepstatin A in a 1.5 ml tube
- Add 1 ml methanol
- Mix until dissolved
- Weigh 1 mg of pepstatin A in a 1.5 ml tube
- 1 mg/ml leupeptin (1 ml)
- Weigh 1 mg of leupeptin in a 1.5 ml tube
- Add 1 ml sterile water
- Mix until dissolved
- Weigh 1 mg of leupeptin in a 1.5 ml tube
- 1 mg/ml aprotinin (1 ml)
- Weigh 1 mg of aprotinin in a 1.5 ml tube
- Add 1 ml sterile water
- Mix until dissolved
- Weigh 1 mg of aprotinin in a 1.5 ml tube
- 50% slurry of Protein A Sepharose CL-4B
- Decant ethanol from beads
- Resuspend beads in 50 mM Tris-HCl to generate a 75% bead slurry
- Centrifuge beads at 1,000 x g for 2 min
- Decant supernatant from beads
- Repeat steps 17b-17d
- Resuspend beads in 50 mM Tris-HCl to generate a 50% bead slurry
- Decant ethanol from beads
- Cdk5 lysis buffer (stock) (50 ml)
1.25 ml of 2 M Tris-HCl (pH 7.5) to a 50 ml conical tube
2.5 ml of 5 M NaCl
100 µl of 0.5 M EDTA (pH 8.0)
Bring final volume to 50 ml using sterile water - Cdk5 lysis buffer (working) (5 ml)
5 ml of Cdk5 lysis buffer (stock) to a 50 ml conical tube
250 µl of 100 mM Na3VO4
25 µl of 1 M NaF
25 µl of 20% NP-40
50 µl of 100 mM PMSF
5 µl of pepstatin
5 µl of leupeptin
5 µl of aprotinin - Cdk5 kinase buffer (stock) (50 ml)
1 ml of 1 M MOPS (pH 7.0) to a 50 ml conical tube
250 µl of 1 M MgCl2
10 µl of 0.5 M EDTA (pH 8.0)
10 µl of 0.5 M EGTA (pH 8.0)
Bring final volume to 50 ml using sterile water - Cdk5 kinase buffer (washes) (5 ml)
5 ml of Cdk5 kinase buffer (stock) to a 15 ml conical tube
50 µl of PMSF - Cdk5 kinase buffer (assay) (1 ml)
1 ml of Cdk5 kinase buffer (stock) to a 15 ml conical tube
50 µl of PMSF
2.5 µl of 10 mM ATP
5 µl of 20 µg/µl histone H1 - MOPS dilution buffer (50 ml)
1 µl of 1 M MOPS buffer to a 50 ml conical tube
100 µl of 0.5 M EDTA (pH 8.0)
50 µl of 10% Brij-35 solution
2.5 ml of 100% glycerol
50 µl of 2-mercaptoethanol
50 mg BSA
Bring final volume to 50 ml using sterile water - 5x reaction buffer (50 ml)
2 µl of 1 M MOPS buffer to a 50 ml conical tube
100 µl of 0.5 M EDTA (pH 8.0) - 50 mM magnesium acetate buffer (MgOAc) (15 ml)
Weigh 0.11 g in a 15 ml conical tube
Add sterile water to bring the final volume to 15 ml - 1x phosphate-buffered saline/0.1% Tween-20 (PBS-T) (500 ml)
50 ml of 10x PBS
1 ml of 100% Tween-20
Bring final volume to 500 ml using sterile water - 5x Laemmli buffer
1 ml glycerol
1 g SDS
1.56 ml 2M Tris-HCl
2.5 ml β-mercaptoethanol
1 ml 0.2% bromophenol blue - 0.2% bromophenol blue
0.02 g bromophenol blue in a 15 ml conical tube
10 ml sterile water
Acknowledgments
This work was supported by NIH/NINDS R01NS093016 and R01NS056097, NMSSRG 4010-A, and an Emory University Research Committee grant (YF). ANB was supported by NIH training grant T32GM008602. This protocol was adapted based on previous studies (Humbert et al., 2000; Yamada et al., 2007; Bankston et al., 2013).
References
- Bankston, A. N., Li, W., Zhang, H., Ku, L., Liu, G., Papa, F., Zhao, L., Bibb, J. A., Cambi, F., Tiwari-Woodruff, S. K. and Feng, Y. (2013). p39, the primary activator for cyclin-dependent kinase 5 (Cdk5) in oligodendroglia, is essential for oligodendroglia differentiation and myelin repair. J Biol Chem 288(25): 18047-18057.
- Asada, A., Yamamoto, N., Gohda, M., Saito, T., Hayashi, N. and Hisanaga, S. (2008). Myristoylation of p39 and p35 is a determinant of cytoplasmic or nuclear localization of active cyclin-dependent kinase 5 complexes. J Neurochem 106(3): 1325-1336.
- He, X., Takahashi, S., Suzuki, H., Hashikawa, T., Kulkarni, A. B., Mikoshiba, K. and Ohshima, T. (2011). Hypomyelination phenotype caused by impaired differentiation of oligodendrocytes in Emx1-cre mediated Cdk5 conditional knockout mice. Neurochem Res 36(7): 1293-1303.
- Hisanaga, S. and Saito, T. (2003). The regulation of cyclin-dependent kinase 5 activity through the metabolism of p35 or p39 Cdk5 activator. Neurosignals 12(4-5): 221-229.
- Humbert, S., Dhavan, R. and Tsai, L. (2000). p39 activates cdk5 in neurons, and is associated with the actin cytoskeleton. J Cell Sci 113 (Pt 6): 975-983.
- Ko, J., Humbert, S., Bronson, R. T., Takahashi, S., Kulkarni, A. B., Li, E. and Tsai, L. H. (2001). p35 and p39 are essential for cyclin-dependent kinase 5 function during neurodevelopment. J Neurosci 21(17): 6758-6771.
- Miyamoto, Y., Yamauchi, J., Chan, J. R., Okada, A., Tomooka, Y., Hisanaga, S. and Tanoue, A. (2007). Cdk5 regulates differentiation of oligodendrocyte precursor cells through the direct phosphorylation of paxillin. J Cell Sci 120(Pt 24): 4355-4366.
- Miyamoto, Y., Yamauchi, J. and Tanoue, A. (2008). Cdk5 phosphorylation of WAVE2 regulates oligodendrocyte precursor cell migration through nonreceptor tyrosine kinase Fyn. J Neurosci 28(33): 8326-8337.
- Ohshima, T., Ward, J. M., Huh, C. G., Longenecker, G., Veeranna, Pant, H. C., Brady, R. O., Martin, L. J. and Kulkarni, A. B. (1996). Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc Natl Acad Sci U S A 93(20): 11173-11178.
- Sato, K., Zhu, Y. S., Saito, T., Yotsumoto, K., Asada, A., Hasegawa, M. and Hisanaga, S. (2007). Regulation of membrane association and kinase activity of Cdk5-p35 by phosphorylation of p35. J Neurosci Res 85(14): 3071-3078.
- Tang, X. M., Strocchi, P. and Cambi, F. (1998). Changes in the activity of cdk2 and cdk5 accompany differentiation of rat primary oligodendrocytes. J Cell Biochem 68(1): 128-137.
- Tsai, L. H., Delalle, I., Caviness, V. S. Jr., Chae, T. and Harlow, E. (1994). p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature 371(6496): 419-23.
- Yamada, M., Saito, T., Sato, Y., Kawai, Y., Sekigawa, A., Hamazumi, Y., Asada, A., Wada, M., Dio, H. and Hisanaga, S. (2007). Cdk5-p39 is a labile complex with the similar substrate specificity to Cdk5-p35. J Neurochem 102: 1477-87
- Yang, Y., Wang, H., Zhang, J., Luo, F., Herrup, K., Bibb, J. A., Lu, R. and Miller, R. H. (2013). Cyclin dependent kinase 5 is required for the normal development of oligodendrocytes and myelin formation. Dev Biol 378(2): 94-106.
- Zheng, M., Leung, C. L. and Liem, R. K. (1998). Region-specific expression of cyclin-dependent kinase 5 (cdk5) and its activators, p35 and p39, in the developing and adult rat central nervous system. J Neurobiol 35(2): 141-159.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Bankston, A. N., Ku, L. and Feng, Y. (2017). Active Cdk5 Immunoprecipitation and Kinase Assay. Bio-protocol 7(13): e2363. DOI: 10.21769/BioProtoc.2363.
- Bankston, A. N., Li, W., Zhang, H., Ku, L., Liu, G., Papa, F., Zhao, L., Bibb, J. A., Cambi, F., Tiwari-Woodruff, S. K. and Feng, Y. (2013). p39, the primary activator for cyclin-dependent kinase 5 (Cdk5) in oligodendroglia, is essential for oligodendroglia differentiation and myelin repair. J Biol Chem 288(25): 18047-18057.
Category
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









