- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Biochemical Analysis of Caspase-8-dependent Proteolysis of IRF3 in Virus-infected Cells
Published: Vol 6, Iss 22, Nov 20, 2016 DOI: 10.21769/BioProtoc.2018 Views: 9803
Reviewed by: Yannick DebingAngela CoronaRosario Gomez-Garcia

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Ub-POD: A Ubiquitin-Specific Proximity-Dependent Labeling Technique to Identify E3 Ubiquitin Ligase Substrates in Human Cells
Urbi Mukhopadhyay [...] Sagar Bhogaraju
Jun 20, 2025 2479 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2473 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3055 Views
Abstract
Interferon regulatory factor 3 (IRF3) is a transcription factor, which is critical for the antiviral response against a wide range of viruses (Hiscott, 2007; Ikushima et al., 2013). It gets activated in virus-infected cells via Toll like receptors (TLRs), RIG-I (retinoic acid inducible gene 1) like receptors (RLRs), cyclic GMP-AMP synthase (cGAS) – stimulator of interferon genes (STING), which are sensors of viral components in the cells (Chattopadhyay and Sen, 2014a; 2014b; Hiscott, 2007). IRF3 is a cytoplasmic protein, upon activation by virally activated sensors it gets phosphorylated, translocated to the nucleus and binds to the interferon-sensitive response element (ISRE) of the gene promoters to induce their transcription (Hiscott, 2007). IRF3 has other functions, including direct stimulation of apoptosis in virus-infected cells. In this pathway, the transcriptional activity of IRF3 is not required (Chattopadhyay et al., 2013b; Chattopadhyay et al., 2016; Chattopadhyay et al., 2010; Chattopadhyay and Sen, 2010; Chattopadhyay et al., 2011). These pathways are negatively regulated by host factors as well as by viruses. Our studies indicate that IRF3 can be proteolytically processed by caspase-8-dependent cleavage (Sears et al., 2011). A specific site in IRF3 is targeted by caspase-8, activated in RNA or DNA virus-infected and dsRNA-stimulated cells (Sears et al., 2011). The direct involvement of caspase-8 was confirmed by in vitro cleavage assay using recombinant proteins and in vivo by virus activated caspase-8. The proteolytic cleavage of IRF3 can be inhibited by chemical inhibition or genetic ablation of caspase-8. The cleavage of IRF3 removes the activated pool of IRF3 and thus can be used as a pro-viral mechanism (Figure 1). Using a C-terminally epitope-tagged human IRF3, we analyzed the cleavage of IRF3 in virus-infected cells. Moreover, we used recombinant proteins in vitro to conclude that IRF3 is a substrate of caspase-8 (Sears et al., 2011). In the current protocol, we have outlined a simple and detailed procedure to biochemically analyze the proteolysis of IRF3 in virus-infected cells and the specific role of caspase-8 in this process. 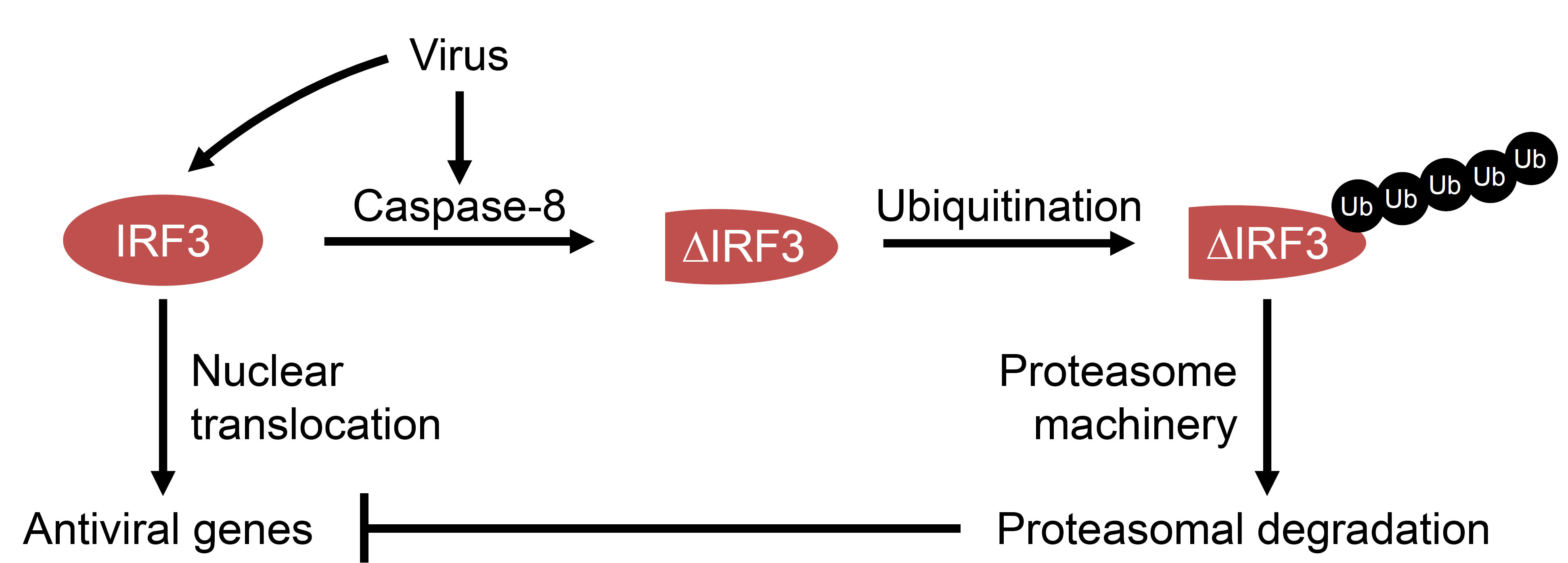
Figure 1. Model for virus-induced caspase-8-dependent proteolysis of IRF3. In virus-infected cells, IRF3 can be proteolytically cleaved by caspase-8, which gets activated during infection. The cleaved IRF3 is subjected to poly-ubiquitination (Ub) and degradation by proteasome machinery. The degradation of IRF3 inhibits dsRNA-induced antiviral gene induction.
Background
IRF3 functions as a transcription factor as well as a pro-apoptotic factor in virus-infected cells. The two properties of IRF3 determine its overall antiviral functions. Our studies have revealed a number of additional regulatory mechanisms of IRF3 for both of its functions. Using genetically modified cells and mice, we have shown how the two functional branches of IRF3 protect against virus infections (Chattopadhyay et al., 2013a; Chattopadhyay et al., 2016; Chattopadhyay et al., 2011). Dysregulated IRF3 activation is undesirable for normal physiological functions and, therefore, negative regulatory mechanisms are in place to control continuous activation of IRF3. In virus-infected cells, IRF3 can be proteolytically cleaved at a specific site by caspase-8, which gets activated by TLR or RLR signaling. The proteolytic cleavage of IRF3 prevents unregulated expression of dsRNA-dependent genes (Sears et al., 2011). We speculate that such mechanisms are used by viruses to promote their replication and to downregulate IRF3 responses. Therefore, the proteolysis of IRF3 represents a critical step, which both the viruses and the host can regulate to their favorable outcomes. This protocol provides a method to biochemically analyze the proteolysis of IRF3 in virus-infected cells.
Materials and Reagents
- Materials
- Autoradiography film (Denville Scientific, catalog number: E3012 )
- Tissue culture plates (Corning)
- 1.5 ml Eppendorf tubes (USA Scientific)
- Centrifuge tubes (Thermo Fisher Scientific)
- 26-gauge needle
- PVDF membranes
- Autoradiography film (Denville Scientific, catalog number: E3012 )
- Cells
- HT1080 fibrosarcoma cells
Note: These cells are maintained in DMEM containing 10% FBS, 100 international units of penicillin, 100 µg/ml streptomycin (complete DMEM). - 1080.10 cells (IRF3 overexpressing HT1080 cells [Elco et al., 2005])
Note: These cells are maintained in complete DMEM containing 1 µg/ml puromycin. - P2.1/IRF3 cells (P2.1 cells expressing C-terminally Flag-tagged human IRF3, Figure 2A)
- These cells were generated by lentiviral transduction of the P2.1 cells with cDNA of Flag-tagged human IRF3. After lentiviral transduction, the cells were selected in puromycin (1 µg/ml)-containing complete medium. Use the selected pool of cells for the experiments.
- HT1080 cells were described previously (Chattopadhyay et al., 2016) and the other cell lines (1080.10 and P2.1/IRF3) were generated in the authors’ laboratory.
- HT1080 fibrosarcoma cells
- Viruses
- Sendai virus (SeV) Cantell strain (Charles River laboratories) – this strain was originally obtained from ATCC (ATCC, catalog number: VR-907TM )
- Adenovirus Ad5 strain (a kind gift from Thomas Shenk, Princeton University, New Jersey)
- Sendai virus (SeV) Cantell strain (Charles River laboratories) – this strain was originally obtained from ATCC (ATCC, catalog number: VR-907TM )
- Reagents
- DMEM (Lerner Research Institute Central Cell Services, catalog number: 11-500p )
- PolyI:polyC (Poly [I:C]) (GE Healthcare, catalog number: 27-4732-01 )
- Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11668030 )
- Opti-MEM I reduced serum medium (Thermo Fisher Scientific, InvitrogenTM, catalog number: 31985070 )
- Phosphate-buffered saline (PBS) (Lerner Research Institute Central Cell Services, catalog number: 123-1000p )
- Recombinant caspase-8 (EMD Millipore, catalog number: CC123 )
- Protein assay dye (Bio-Rad Laboratories, catalog number: 5000006 )
- 10x SDS-PAGE running buffer (AMRESCO, catalog number: 0783 )
- Antibodies:
anti-Flag (used at 1:2,500) (Sigma-Aldrich, catalog number: F3290 )
anti-IRF3 (dilution used at 1:10,000) (Active Motif, catalog number: 39033 )
anti-actin (used at 1:10,000 ) (Sigma-Aldrich, catalog number: A5441 )
HRP-conjugated secondary antibodies (used at 1:5,000) (Rockland Immunochemicals, catalog numbers: 611-103-121 , 611-103-122 ) - Fetal bovine serum (FBS) (Atlanta Biologicals, catalog number: S11550 )
- Proteasome inhibitor MG132 (EMD Millipore, catalog number: 474790 )
- Phosphatase inhibitor (PhosSTOP) (Roche Diagnostics, catalog number: 04906837001 )
- cOmplete EDTA-free protease inhibitor (Roche Diagnostics, catalog number: 11873580001 )
- Caspase inhibitors:
z-VAD (MBL, catalog number: 4800-520 )
z-WEHD (MBL, catalog number: BV-1100-3 )
z-IETD (MBL, catalog number: 4805-510 ) - Anti-Flag-agarose beads (Sigma-Aldrich, catalog number: A2220 )
- FLAG peptide (Sigma-Aldrich, catalog number: F3290 )
- SDS-PAGE loading buffer (Laemelli) (Bio-Rad Laboratories, catalog number: 161-0737 )
- Tris buffered saline with Tween-20 (TBS-T) (AMRESCO, catalog number: K873 )
- Nonfat dry milk (Bio-Rad Laboratories, catalog number: 1706404XTU )
- PierceTM ECL2 (Thermo Fisher Scientific, Fisher Scientific, catalog number: PI80196 )
- Tris buffer (pH 7.4) (Affymetrix, catalog number: 22639 )
- Sodium chloride (NaCl) (Affymetrix, catalog number: 75888 )
- Triton X-100 (Sigma-Aldrich, catalog number: T9284 )
- Sodium orthovanadate (NaVO4) (Sigma-Aldrich, catalog number: S6508 )
- Sodium fluoride (NaF) (Sigma-Aldrich, catalog number: S7920 )
- β-glycerophosphate (Sigma-Aldrich, catalog number: G9422 )
- Sodium pyrophosphate (Na2H2P2O7) (Sigma-Aldrich, catalog number: P8135 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- CHAPS hydrate (Sigma-Aldrich, catalog number: C3023 )
- EDTA (Affymetrix, catalog number: 15694 )
- Glycerol (Affymetrix, catalog number: 16374 )
- 1,4-dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 10197777001 )
- 10x transfer buffer (AMRESCO, catalog number: 0307 )
- Cell lysis buffer (see Recipes)
- Caspase-8 in vitro cleavage assay buffer (see Recipes)
- DMEM (Lerner Research Institute Central Cell Services, catalog number: 11-500p )
Equipment
- Tissue culture incubator (at 37 °C with 5% CO2) (Thermo Fisher Scientific)
- Biosafety cabinet
- Table top centrifuge (Eppendorf)
- Water bath (Benchmark)
- Heating block (at 95 °C) (Benchmark)
- SDS-PAGE and transfer apparatus (Bio-Rad Laboratories, model: Mini PROTEAN-II Cell )
Note: This equipment is no longer available at manufacturer. - Rocker (Benchmark)
- Rotator (Labnet)
- Refrigerator
- Autoradiography film processor (Kodak)
- Spectrophotometer
- Vortex (Thermo Fisher Scientific)
Software
- ImageJ (freely available from National Institutes of Health, https://imagej.nih.gov/ij/)
Procedure
- Proteolysis of IRF3 in virus-infected cells (Figure 2B)
- For virus-induced IRF3 cleavage, the HT1080 (expressing endogenous IRF3) or 1080.10 (expressing higher levels of human IRF3) cells are used.
- Seed 500,000 cells/well (in 6-well plates) in complete DMEM for virus infection ([Sendai virus, multiplicity of infection, moi: 10] or [Adenovirus, moi: 10]) or in antibiotic-free DMEM for poly (I:C) transfection as described below. After 24 h, use these cells for induction of proteolysis of IRF3 by virus infection or poly (I:C) transfection as described below.
- Induction of IRF3 proteolysis
- Virus infection: Wash the cells in serum-free DMEM and infect with SeV (moi: 10) in 2% serum containing DMEM for 1 h, with occasional gentle shaking. At the end of infection, wash the cells with PBS and place in 10% serum containing DMEM. For Ad5 infection (moi: 10), use the above infection protocol in serum-free DMEM.
- Poly (I:C) transfection: Transfect with poly (I:C) (2 µg/ml) using Lipofectamine 2000 (at 1:2 ratio of poly [I:C] and Lipofectamine 2000 in opti-MEM).
- Virus infection: Wash the cells in serum-free DMEM and infect with SeV (moi: 10) in 2% serum containing DMEM for 1 h, with occasional gentle shaking. At the end of infection, wash the cells with PBS and place in 10% serum containing DMEM. For Ad5 infection (moi: 10), use the above infection protocol in serum-free DMEM.
- At the desired time post infection (0-12 h for SeV infection, 0-96 h for Ad5 infection and 8 h for poly (I:C) transfection (Sears et al., 2011), harvest the cells in cold PBS and lyse in 30 µl of lysis buffer by incubating on ice for 30 min with occasional vortexing. Centrifuge at 12,000 x g for 15 min and collect the soluble cell lysates.
- Measure protein concentrations by adding 1 µl of the cell lysates in 1 ml of pre-diluted (1:5) Bio-Rad protein assay dye and measuring the absorbance at 595 nm after 5 min of incubation at room temperature.
- Analyze equal amounts (20 µg) of protein from the cell lysates by SDS-PAGE followed by Western blot with anti-Flag antibody to detect the full length and cleaved forms of IRF3.
- For virus-induced IRF3 cleavage, the HT1080 (expressing endogenous IRF3) or 1080.10 (expressing higher levels of human IRF3) cells are used.
- Involvement of caspase-8 in proteolysis of IRF3 (Figure 2C)
- Seed 500,000 P2.1/IRF3 cells in 6-well plate in complete DMEM and for poly (I:C) transfection, seed the cells in antibiotic-free DMEM (in 10% FBS).
- After 24 h, pre-treat the cells with various inhibitors of caspases (z-VAD: pan caspase inhibitor, z-WEHD: caspase-1 inhibitor and z-IETD: caspase-8 inhibitor) at 10 µM for 1 h.
- Induce proteolysis of IRF3 by adding poly (I:C) to the culture medium (100 µg/ml, TLR3 activation, T in Figure 2C) or transfection of poly (I:C) using Lipofectamine 2000 (2 µg/ml or poly [I:C], RLR activation, R in Figure 2C).
- Harvest cells at 8 h post poly (I:C) treatment in cold PBS and lyse in 30 µl of lysis buffer by incubating on ice for 30 min, with occasional vortexing. Centrifuge at 12,000 x g for 15 min and collect the soluble cell lysates.
- Measure protein concentrations by adding 1 µl of the cell lysates in 1 ml of pre-diluted (1:5) Bio-Rad protein assay dye and measuring the absorbance at 595 nm after 5 min of incubation at room temperature.
- Analyze equal amounts (20 µg) of protein from cell lysates by SDS-PAGE followed by Western blot with anti-Flag antibody (at 1:2,500) to detect the full length and cleaved forms of IRF3.
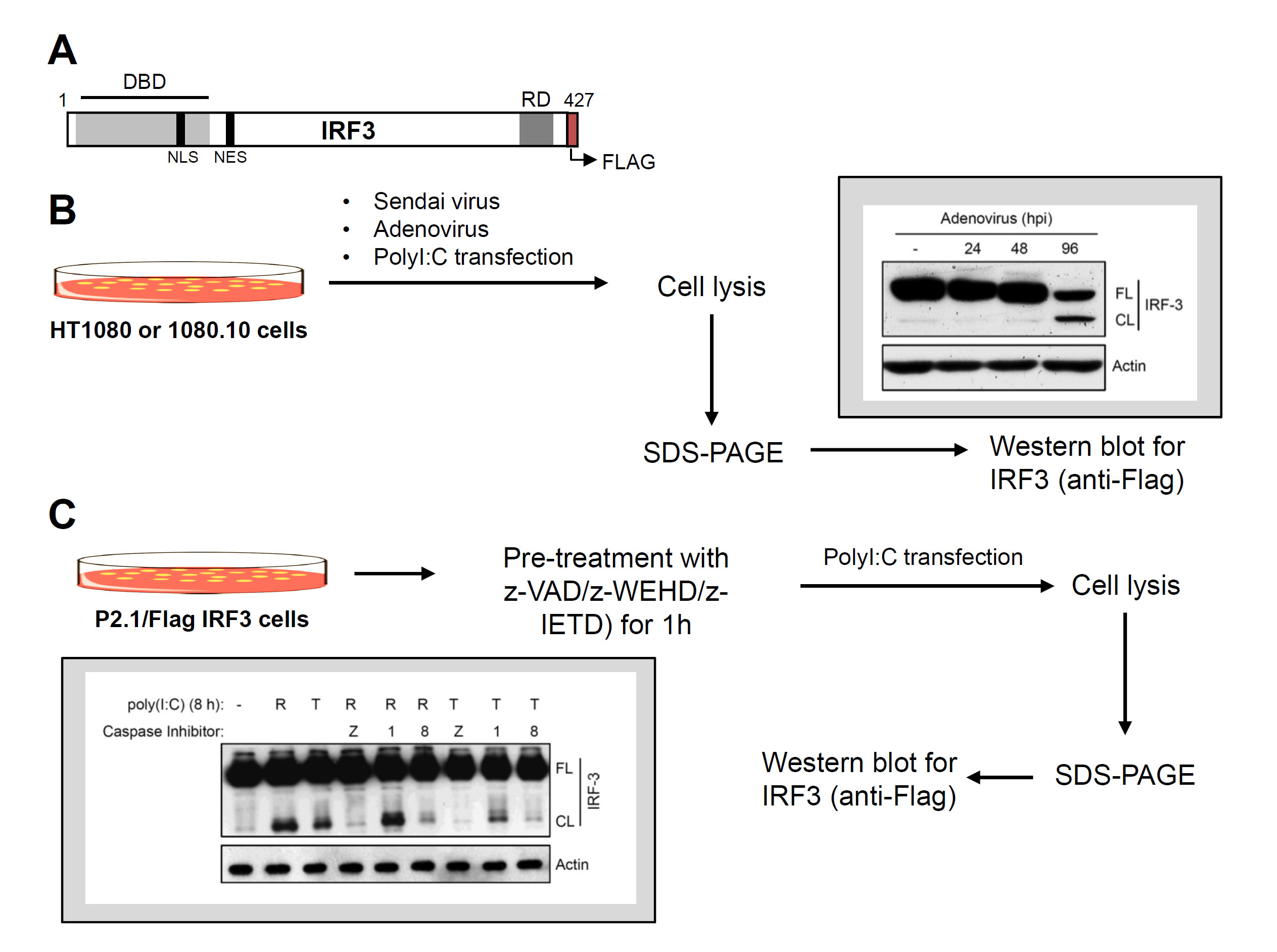
Figure 2. Virus-induced proteolysis of IRF3 and caspase-8 involvement. A. C-terminally Flag-tagged human IRF3 and its functional domains, DBD, DNA binding domain, NLS, nuclear localization signal, NES, nuclear export signal, RD, regulatory domain. B. Flow chart of virus infection and dsRNA (poly [I:C])-induced proteolysis of IRF3 (inset shows a representative result of virus-induced IRF3 cleavage). C. Flow chart of proteolysis of IRF3 in the absence or the presence of various caspase inhibitors (as indicated) (inset shows a representative result).
- Seed 500,000 P2.1/IRF3 cells in 6-well plate in complete DMEM and for poly (I:C) transfection, seed the cells in antibiotic-free DMEM (in 10% FBS).
- In vitro cleavage of IRF3 by caspase-8 (Figure 3)
- Purification of Flag-IRF3
- Seed P2.1/IRF3 cells (106 cells/plate) in complete DMEM in twenty-five 10 cm tissue culture plates. When the plates are confluent, harvest the cells in cold PBS, centrifuge at 9,000 x g to collect the cell pellet. Lyse the cells in 2 ml of lysis buffer, incubate on ice for 30 min and centrifuge at 12,000 x g to clear the lysate. Collect the supernatants as cell lysate to purify IRF3.
- Incubate the cell lysate with anti-Flag-agarose beads (100 µl bead volume) for 16 h at 4 °C by placing on a rotator.
- Centrifuge at 2,000 x g for 1 min, discard the supernatant and wash the beads three times with 1 ml of cell lysis buffer.
- Elute the bound IRF3 from the beads using Flag peptide (at a final concentration of 25 µg/ml) by placing the washed beads in elution buffer (Flag peptide in the cell lysis buffer) on a rotator. Confirm by Western blot that the eluted proteins contain the full-length IRF3 protein by using anti-Flag and anti-IRF3 antibodies.
- Use the eluted IRF3 as a substrate for in vitro proteolysis.
- Seed P2.1/IRF3 cells (106 cells/plate) in complete DMEM in twenty-five 10 cm tissue culture plates. When the plates are confluent, harvest the cells in cold PBS, centrifuge at 9,000 x g to collect the cell pellet. Lyse the cells in 2 ml of lysis buffer, incubate on ice for 30 min and centrifuge at 12,000 x g to clear the lysate. Collect the supernatants as cell lysate to purify IRF3.
- Incubate the purified Flag-IRF3 with various amounts (0.01-1 units) of recombinant active caspase-8 in the cleavage assay buffer at 37 °C for increasing lengths of time (0-3 h) in 50 µl of total reaction volume in 1.5 ml Eppendorf tubes.
- Terminate the reaction by adding SDS-PAGE loading buffer.
- Analyze equal protein amount from the reaction mixture by SDS-PAGE followed by Western blot using anti-Flag antibody.
- See Figure 3 for a representative result of in vitro IRF3 cleavage.
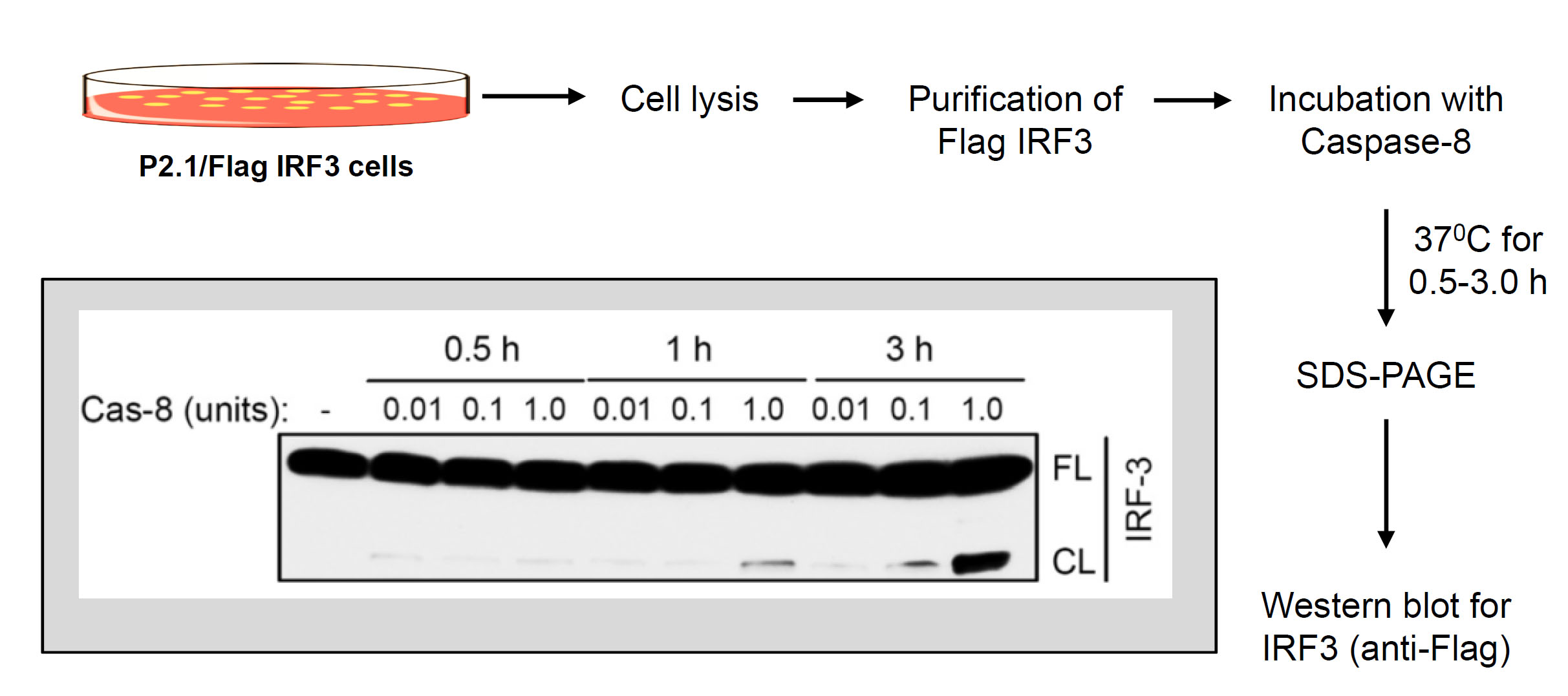
Figure 3. In vitro proteolysis of IRF3 by recombinant caspase-8. Flow chart of in vitro proteolysis of IRF3 by recombinant caspase-8 (inset shows a representative result of caspase-8-induced in vitro cleavage of IRF3).
- Purification of Flag-IRF3
- SDS-PAGE and Western blot
SDS-PAGE and Western blot procedures were followed from the standard protocols described elsewhere (Towbin et al., 1979; Renart et al., 1979). Briefly, follow the steps below:- Load equal amounts of protein from the cell lysates or in vitro reactions on 10% SDS gels and electrophorese at 100 V for 2 h to separate the proteins.
- At the end of the electrophoresis, transfer the separated proteins to the PVDF membranes using a Bio-Rad wet transfer apparatus at 300 mA for 1 h.
- Block the transferred proteins on the membranes in TBS-T containing 5% nonfat dry milk (blocking buffer) for 1 h at room temperature on a rocker.
- Incubate the membranes with primary antibodies diluted (as described in the procedures) in the blocking buffer for 16 h at 4 °C on a rocker.
- Wash the membranes three times with TBS-T and incubate with the HRP-conjugated secondary antibodies, diluted in blocking buffer, for 1 h at room temperature on a rocker.
- Wash the membranes with TBS-T for three times and incubate in ECL2 reagents and develop on autoradiography films using automated the film processor.
- Load equal amounts of protein from the cell lysates or in vitro reactions on 10% SDS gels and electrophorese at 100 V for 2 h to separate the proteins.
- Quantification of IRF3 cleavage
Use the autoradiograph of the Western blots to analyze IRF3 cleavage by ImageJ computer program. Using ImageJ, measure the band intensities of the full length (higher molecular weight band, ~55 kDa) and the cleaved (lower molecular weight band, ~45 kDa) forms of IRF3. Percentage cleavage of IRF3 is calculated as:
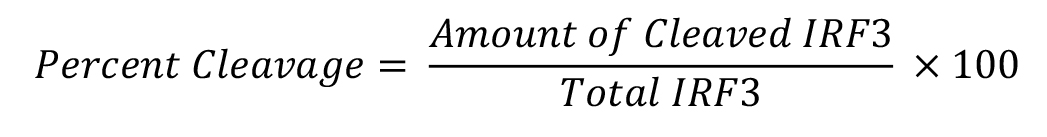
Percent cleavage of IRF3 is presented as a representative of multiple independent experiments. The quantitative estimation of IRF3 cleavage has been reported previously (Sears et al., 2011).
Notes
Special care should be taken to separate the cleaved (~45 kDa), and the full length (~55 kDa), forms of IRF3, by electrophoresing the SDS gel (10%) at 100 V for longer time (when the protein ladders indicate clear separation of the desired sizes).
Recipes
- Cell lysis buffer
50 mM Tris buffer, pH 7.4
150 mM NaCl
0.1% Triton X-100
1 mM sodium orthovanadate
10 mM sodium fluoride
10 mM glycerophosphate
5 mM sodium pyrophosphate - Caspase-8 in vitro cleavage assay buffer
50 mM HEPES
50 mM NaCl
0.1% CHAPS
10 mM EDTA
5% glycerol
10 mM DTT - Poly (I:C) preparation
Re-suspend poly (I:C) in PBS (at a final concentration of 1 mg/ml)
Pass through a 26-gauge needle several times to shear
Acknowledgments
Our work is supported by American Heart Association Scientist Development grants 15SDG25090212 (SC) and 15SDG2308025 (RC), and the University of Toledo College of Medicine and Life Sciences startup funds (SC). The protocol and the representative results were adapted from our previous work (Sears et al., 2011).
References
- Chattopadhyay, S., Fensterl, V., Zhang, Y., Veleeparambil, M., Wetzel, J. L. and Sen, G. C. (2013a). Inhibition of viral pathogenesis and promotion of the septic shock response to bacterial infection by IRF-3 are regulated by the acetylation and phosphorylation of its coactivators. MBio 4(2).
- Chattopadhyay, S., Fensterl, V., Zhang, Y., Veleeparambil, M., Yamashita, M. and Sen, G. C. (2013b). Role of interferon regulatory actor 3-mediated apoptosis in the establishment and maintenance of persistent infection by Sendai virus. J Virol 87(1): 16-24.
- Chattopadhyay, S., Kuzmanovic, T., Zhang, Y., Wetzel, J. L. and Sen, G. C. (2016). Ubiquitination of the transcription factor IRF-3 activates RIPA, the apoptotic pathway that protects mice from viral pathogenesis. Immunity 44(5): 1151-1161.
- Chattopadhyay, S., Marques, J.T., Yamashita, M., Peters, K.L., Smith, K., Desai, A., Williams, B.R. and Sen, G.C. (2010). Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J 29: 1762-1773.
- Chattopadhyay, S. and Sen, G. C. (2010). IRF-3 and Bax: a deadly affair. Cell Cycle 9(13): 2479-2480.
- Chattopadhyay, S. and Sen, G. C. (2014a). dsRNA-activation of TLR3 and RLR signaling: gene induction-dependent and independent effects. J Interferon Cytokine Res 34(6): 427-436.
- Chattopadhyay, S. and Sen, G. C. (2014b). Tyrosine phosphorylation in Toll-like receptor signaling. Cytokine Growth Factor Rev 25(5): 533-541.
- Chattopadhyay, S., Yamashita, M., Zhang, Y. and Sen, G. C. (2011). The IRF-3/Bax-mediated apoptotic pathway, activated by viral cytoplasmic RNA and DNA, inhibits virus replication. J Virol 85(8): 3708-3716.
- Elco, C. P., Guenther, J. M., Williams, B. R. and Sen, G. C. (2005). Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not toll-like receptor 3. J Virol 79(7): 3920-3929.
- Hiscott, J. (2007). Triggering the innate antiviral response through IRF-3 activation. J Biol Chem 282: 15325-15329.
- Ikushima, H., Negishi, H. and Taniguchi, T. (2013). The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb Symp Quant Biol 78: 105-116.
- Renart, J., Reiser, J. and Stark, G. R. (1979). Transfer of proteins from gels to diazobenzyloxymethyl-paper and detection with antisera: a method for studying antibody specificity and antigen structure. Proc Natl Acad Sci U S A 76(7): 3116-3120.
- Sears, N., Sen, G. C., Stark, G. R. and Chattopadhyay, S. (2011). Caspase-8-mediated cleavage inhibits IRF-3 protein by facilitating its proteasome-mediated degradation. J Biol Chem 286(38): 33037-33044.
- Towbin, H., Staehelin, T. and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76(9): 4350-4354.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Subramanian, G., Pan, K., Chakravarti, R. and Chattopadhyay, S. (2016). Biochemical Analysis of Caspase-8-dependent Proteolysis of IRF3 in Virus-infected Cells. Bio-protocol 6(22): e2018. DOI: 10.21769/BioProtoc.2018.
- Sears, N., Sen, G. C., Stark, G. R. and Chattopadhyay, S. (2011). Caspase-8-mediated cleavage inhibits IRF-3 protein by facilitating its proteasome-mediated degradation. J Biol Chem 286(38): 33037-33044.
Category
Microbiology > Microbe-host interactions > Virus
Molecular Biology > Protein > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











