- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RAB21 Activity Assay Using GST-fused APPL1
Published: Vol 6, Iss 4, Feb 20, 2016 DOI: 10.21769/BioProtoc.1738 Views: 8607
Reviewed by: Ralph BottcherVaibhav B ShahAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurement of the Activity of Wildtype and Disease-Causing ALPK1 Mutants in Transfected Cells With a 96-Well Format NF-κB/AP-1 Reporter Assay
Tom Snelling
Nov 20, 2024 1609 Views

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2282 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views
Abstract
The Rab family of small GTPases are essential regulators of membrane trafficking events. As with other small GTPase families, Rab GTPases cycle between an inactive GDP- bound state and an active GTP-bound state. Guanine nucleotide exchange factors (GEFs) promote Rab activation with the exchange of bound GDP for GTP, while GTPase-activating proteins (GAPs) regulate Rab inactivation with GTP hydrolysis. Numerous methods have been established to monitor the activation status of Rab GTPases. Of those, FRET-based methods are used to identify when and where a Rab GTPase is activated in cells. Unfortunately, the generation of such probes is complex, and only a limited number of Rabs have been probed this way. Biochemical purification of activated Rabs from cell or tissue extracts is easily achievable through the use of a known Rab effector domain to pull down a specific GTP-bound Rab form. Although this method is not ideal for detailed subcellular localization, it can offer temporal resolution of Rab activity. The identification of a growing number of specific effectors now allows tests for activation levels of many Rab GTPases in specific conditions. Here, we described an affinity purification approach using GST fused APPL1 (a known RAB21 effector) to test RAB21 activation in mammalian cells. This method was successfully used to assay changes in RAB21 activation status under nutrient rich versus starved conditions and to test the requirement of the MTMR13 RAB21 GEF in this process.
Keywords: Membrane traffickingMaterials and Reagents
- 50 ml conical tube (Sarstedt AG & CO, catalog number: 62.547.004 )
- 100 mm Tissue Culture Dish (Corning, catalog number: 430167 )
- 0.22 µm filtering unit (Genesee Scientific Corporation, catalog number: 25-227 )
- Cell lifter (Corning, catalog number: 3008 )
- Escherichia coli BL21 (New England Biolabs, catalog number: C2530H )
- pGEX-5X-3 (GE Healthcare, catalog number: 27-4586-01 )
- pGEX5X3-Happl1 (aa 5-419) (self-made) (Jean et al., 2015)
- pAcEGFP-C1 vector (Clontech, catalog number: 632470 )
- pEGFP-C1:RAB21 wild type [human RAB21 cloned in pAcEGFP-C1 (this construct was used to generate the stable HeLa M cell line)] (self-made) (Jean et al., 2015)
- EGFP:RAB21wt (wildtype) stably-transfected HeLa M cells (self-made) (Jean et al., 2015)
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Biopioneer, catalog number: c0012 )
- Ampicillin Sodium Salt (Crystalline Powder) (Thermo Fisher Scientific, catalog number: BP176025 )
- BD BactoTM Tryptone (Thermo Fisher Scientific, catalog number: DF0123173 )
- Yeast Extract (Thermo Fisher Scientific, catalog number: 212750 )
- Glutathione Sepharose 4B beads (GE Healthcare, catalog number: 17-0756-01 )
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P8340-5 )
- Fetal Bovine Serum (Sigma-Aldrich, catalog number: F2442-500 ml )
- Penicillin-Streptomycin solution (Life Technologies, catalog number: 15140-122 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 15140-122”. - Trypsin-EDTA (0.25%), phenol red (Life Technologies, catalog number: 25200-056 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 25200-056”. - 0.4% Trypan-Blue (Life Technologies, InvitrogenTM, catalog number: 15250-061 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 15250-061”. - Guanosine 5’-triphosphate sodium salt hydrate (Sigma-Aldrich, catalog number: G8877-25 mg )
- 0.5 M liquid EDTA (Cell grow, catalog number: 45001-122 )
Note: Currently, it is “VWR International, catalog number: 45001-122 ”. - Earl’s Balanced Salt Solution (with sodium bicarbonate, without phenol red liquid, sterile-filtered) (Sigma-Aldrich, catalog number: E3024-500 ml )
- Dithiothreitol (VWR International, catalog number: IB21040 )
- HEPES (Thermo Fisher Scientific, Biotech, catalog number: BP310-100 )
- IGEPAL CA-630 (Sigma-Aldrich, catalog number: I8896-100 ml )
- Glycerol (Thermo Fisher Scientific, catalog number: BP229-1 )
- Magnesium Chloride Hexahydrate (MgCl2.6H2O) (Thermo Fisher Scientific, catalog number: BP214-500 )
- Sodium orthovanadate (Na3VO4) (Sigma-Aldrich, catalog number: S6508-10 g )
- Triton X-100 (Sigma-Aldrich, catalog number: X100-500 ml )
- Sodium Chloride (NaCl) (Thermo Fisher Scientific, catalog number: S671-3 )
- Potassium Chloride (KCl) (VWR International, catalog number: BDH9258-500 g )
- Sodium Phosphate Dibasic Anhydrous (Na2HPO4) (Thermo Fisher Scientific, catalog number: S374500 )
- Potassium Phosphate Monobasic (KH2PO4) (Thermo Fisher Scientific, catalog number: P285-S500 )
- Dubelcco Modified Eagle Medium with High glucose with 4 mM L-Glutamine and sodium pyruvate (GE Healthcare, HycloneTM, catalog number: SH30243.FS ) (see Recipes for complete DMEM)
- Anti-GFP (B2) antibody (Santa Cruz Biotechnology, catalog number: sc-9996 )
- Luria Broth (see Recipes)
- LB/ampicillin (see Recipes)
- Complete DMEM (see Recipes)
- 1x Phosphate Buffer Saline (PBS) (see Recipes)
- 1 M HEPES (pH 7.5) (see Recipes)
- 5 M NaCl (see Recipes)
- 1 M MgCl2 (see Recipes)
- 0.2 M Sodium Orthovanadate (see Recipes)
- 25 mM GTP (see Recipes)
- Modified Magnesium Lysis Buffer (MLB) (see Recipes)
Equipment
- Pipettes 10 μl, 100 μl and 1,000 μl (Eppendorf, catalog numbers: 3120000020 , 3120000046 and 31200000623 )
- Sonifier® cell disrupters (VWR International, catalog number: CA33995-320 )
- 37 °C shaker [similar to New Brunswick Excella E25 (Eppendorf, New Brunswick Scientific, catalog number: M1353-0002 )]
- 30 °C shaker [similar to New Brunswick Excella E25 (Eppendorf, New Brunswick Scientific, catalog number: M1353-0002 )]
- Tissue culture biosafety cabinet [similar to Thermo Scientific 1300 Series Class II (Thermo Fisher Scientific, catalog number: 1323 )]
- Hemacytometer (VWR International, catalog number: 100498-470 )
- Centrifuge (Eppendorf, catalog number: 5804R )
- Table top centrifuge (Eppendorf, catalog number: 5415D )
- Rotating wheel (Barnstead Thermolyne Lab Quake Shaker Rotisserie)
- Sonicator, Branson sonifier S250A Analog Ultrasonic Cell Disruptor (Thermo Fisher Scientific, catalog number: 22309782 )
Procedure
- GST:APPL1 (adaptor protein containing PH domain, PTB domain and Leucine zipper motif) bacterial expression
- Using sterile technique, strike an LB-ampicillin petri dish of E. coli BL21 transformed with pGEX5X3-hAPPL1 (aa 5-419), and incubate at 37 °C overnight (o/n). Plates should be used within 1
week to have maximal protein expression.
- Inoculate 100 ml of LB/ampicillin with one colony from the stroke plate. Grow under constant
agitation at 37 °C o/n.
- Centrifuge the o/n culture at 4,000 x g for 10 min.
- Resuspend the pellet in 50 ml of LB/ampicillin, and mix with 950 ml of LB/ampicillin.
- Grow the culture at 30 °C with agitation until an OD600 of 0.8 is reached, usually by 90 min, then induce the expression of GST:hAPPL1 (5-419) by adding a final concentration of 1 mM
IPTG. Incubate for 3 h at 30 °C under constant agitation.
- After induction, pellet the cells by centrifugation at 4,000 x g for 10 min at 4 °C. Wash the pellet once with 50 ml of 1x PBS. Pellet the cells again by centrifugation. Remove the PBS, and freeze the pellet at -80 °C or move on with the purification.
- Using sterile technique, strike an LB-ampicillin petri dish of E. coli BL21 transformed with pGEX5X3-hAPPL1 (aa 5-419), and incubate at 37 °C overnight (o/n). Plates should be used within 1
week to have maximal protein expression.
- GST:APPL1 (5-419) purification
- Resuspend bacteria in 25 ml ice-cold 1x PBS containing 1x Protease inhibitor cocktail and 1 mM DTT, and transfer the cell suspension into a 50 ml conical tube.
- Sonicate four times for 45 sec each time at 100% duty cycle and at an output of 4 (using a Branson sonifier). Make sure to cool down the sample between each sonication.
- Add 2.5 ml of a 10% Triton X-100/PBS solution to the bacterial lysate for a final Triton concentration of 1%.
- Sonicate as above 1 more time.
- To pellet bacterial debris, centrifuge for 20 min at 12,000 x g at 4 °C.
- While the centrifugation is ongoing, prepare Glutathione Sepharose 4B beads:
- Pipet 800 μl of glutathione beads into an Eppendorf tube.
- Centrifuge beads in a table top centrifuge for 30 sec at 12,000 x g at room
temperature.
- Remove supernatant, and add 1 ml of 1x sterile PBS. Resuspend Glutathione Sepharose 4B beads.
- Repeat steps B6b-c for two more times.
- After the last wash, resuspend Glutathione Sepharose 4B beads in 500 μl of 1x PBS.
- Transfer the resuspended Glutathione Sepharose 4B beads into a 50 ml conical tube.
- Pipet 800 μl of glutathione beads into an Eppendorf tube.
- After centrifugation of the bacterial lysate, pour the supernatant into the 50 ml conical tube containing the prepared Glutathione Sepharose 4B beads. Avoid transferring any E. coli crude.
- To avoid degradation of GST:APPL1 during the room temperature incubation, add 250 μl of 100x Protease inhibitor cocktail to the slurry.
- Incubate with slight agitation 20 min at room temperature.
- Centrifuge the slurry in a tabletop centrifuge at 4,000 x g for 3 min at 4 °C.
- Remove the supernatant and resuspend Glutathione Sepharose 4B beads in 45 ml of 1x ice-cold PBS.
- Repeat steps B10-11 four more times.
- After the last wash, remove most of the 1x PBS. Transfer the slurry to an Eppendorf tube, and assess the quality and quantity of the purified protein through regular SDS-PAGE electrophoresis followed by Coomassie staining (Figure 1). We suggest to use increasing amounts of the GST:APPL1 slurry to assess purity (1 µl, 5 µl, 10 µl and 20 µl of the slurry will cover a large range of protein concentration). Serial BSA dilution series can be used for quantification.
- Store beads at 4 °C. GST:APPL1 beads were used in the same month after their preparation, and experiments were successfully reeated over a month. However, we haven't tested longer storage time.
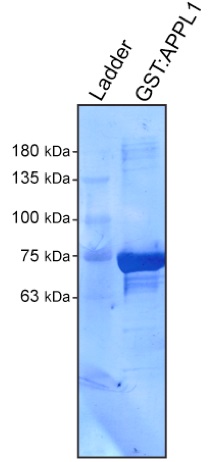
Figure 1. Coomasie stained PVDF membrane of purified GST:APPL1
- Resuspend bacteria in 25 ml ice-cold 1x PBS containing 1x Protease inhibitor cocktail and 1 mM DTT, and transfer the cell suspension into a 50 ml conical tube.
- GFP:RAB21wt HeLa cell culture and autophagy induction
- Remove media from 100 mm stock GFP:RAB21wt cell culture plate, and wash with 5 ml of 1x PBS.
- Remove PBS, add 1 ml of 0.25% trypsin-EDTA, then incubate at 37 °C for 3 min.
- Wash cells off the plate with 10 ml of complete DMEM.
- Pipet 20 μl of the cell suspension, and add to 100 μl of Trypan-Blue solution.
- Pipet 10 μl of this cell/Trypan-Blue dilution into a hemacytometer chamber.
- Count viable cells using the hemacytometer on a standard microscope. Viable cells will be Trypan-Blue negative.
- Add a total of 5 x 105 cells per 100 mm dish (1 dish is required per time point).
- Bring to 8 ml per dish with complete DMEM, and move to 37 °C incubator overnight (o/n).
- To elicit starvation-induced autophagy, wash the cells 1x with 5 ml of pre-warmed EBSS.
- Remove the EBSS, and add 8 ml of pre-warmed EBSS. Incubate for the desired amount of time (typical time points for GFP:Rab21wt HeLa cells, were 0, 5, 15, 30 and 60 min) at 37 °C. To ease subsequent manipulations, start with the longest time points first so that all the different timepoints can be processed concurrently.
- Remove media from 100 mm stock GFP:RAB21wt cell culture plate, and wash with 5 ml of 1x PBS.
- RAB21 activity assay
- Transfer cells on ice, and wash cells twice with ice cold 1x PBS.
- Remove PBS, and lyse cells in 800 μl of ice-cold modified MLB buffer. Scrape cells off the
plate using a cell lifter, and transfer cell lysate to a 1.5 ml Eppendorf tube.
- Incubate cell lysate for 20 min on ice.
- Centrifuge cell lysate 10 min at 16,100 x g at 4 °C.
- While centrifugation is in progress, prepare the GST:APPL1 beads for the experiment:
- Resuspend GST:APPL1 beads, and pipet the appropriate volume into a new 1.5 ml Eppendorf tube. 4 μg of GST:APPL1 is required per time point, thus if 5 time points are tested and the GST:APPL1 is at a concentration of 160 ng/μl, then 25 μl of GST:APPL1 is required per tube.
- Centrifuge the GST:APPL1 beads 30 sec at 11,000 x g at 4 °C.
- Remove supernatant, and add 1 ml of modified MLB buffer.
- Repeat steps D5b-c two more times.
- Remove supernatant.
- Resuspend GST:APPL1 beads, and pipet the appropriate volume into a new 1.5 ml Eppendorf tube. 4 μg of GST:APPL1 is required per time point, thus if 5 time points are tested and the GST:APPL1 is at a concentration of 160 ng/μl, then 25 μl of GST:APPL1 is required per tube.
- For each time point, add 700 μl of the GFP:RAB21wt HeLa cell supernatant to the prepared beads. If different cell lines are compared, the total amount of protein for each cell line lysate must be measured and adjusted in order to have equal amounts of protein incubated with GST:APPL1.
- Incubate on a rotating wheel for 1 h at 4 °C.
- Centrifuge each tube 30 sec at 11,000 x g at room temperature
- Remove supernatants, and add 1 ml of ice-cold modified MLB buffer.
- Repeat steps 8 and 9 three times (for a total of four washes).
- Elute bound proteins from beads by adding 30 μl of 2x SDS-PAGE loading buffer, and heat
at 95 °C for 5 min.
- Analyze the result through western blot analysis with antibodies against GFP (Figure 2).
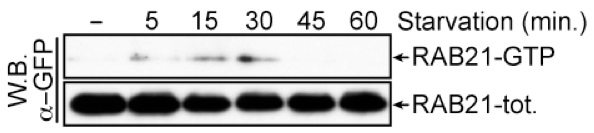
Figure 2. Typical result of a GFP:RAB21 activity assay. Upon nutrient deprivation, GFP:RAB21 is transiently activated. GFP:RAB21-GTP was detected by GST:APPL1 pull-down. Total RAB21 represents RAB21 in cell lysates. Both RAB21-total and RAB21-GTP were detected with an anti-GFP western blot.
- Transfer cells on ice, and wash cells twice with ice cold 1x PBS.
Recipes
- Luria Broth (LB)
Mix 10 g of Tryptone, 5 g of yeast extract and 10 g of NaCl
Add 1 L of water
Mix well and autoclave for 30 min at 121 °C - LB/ampicillin
To 1 L of LB, add 1 ml of ampicillin stock solution (50 mg/ml) - Complete DMEM
Remove 55 ml of DMEM from the purchased 500 ml HyClone bottle
Add 50 ml of fetal bovine serum
Add 5 ml of Penicillin/Streptomycin solution
Filter sterilize using filtering device - 1x phosphate buffer saline
(PBS)
Mix 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4 and 0.24 g of KH2PO4
Add 900 ml of dH2O
Adjust pH to 7.4
Adjust final volume to 1,000 ml
Sterilized by autoclaving at 121 °C for 20 min - 1 M HEPES (pH 7.5)
Mix 238.3 g of HEPES in 900 ml of water
Adjust pH to 7.5 with NaOH
Adjust to 1,000 ml
Sterilized by autoclaving at 121 °C for 20 min - 5 M NaCl
Dissolve 292 g of NaCl in 800 ml of water
Adjust to a final volume of 1,000 ml
Sterilized by autoclaving at 121 °C for 20 min - 1 M MgCl2
Dissolve 203 g of MgCl2.6H2O in 700 ml of water
Adjust to a final volume of 1,000 ml
Sterilized by autoclaving at 121 °C for 20 min - 0.2 M sodium orthovanadate
Dissolve 3.68 g of Na3VO4 in 100 ml of water
Vortex and aliquot for storage
Stored at -20 °C - 25 mM GTP
Add 0.13 g of GTP to 10 ml of water
Filter using a 0.2 μm filter and aliquot for storage
Stored at -20 °C - Modified MLB buffer
Add to 50 ml of water
2.5 ml of 1 M HEPES (pH 7.5) stock solution
3 ml of 5 M NaCl stock solution
2 ml of 1 M MgCl2
10 ml of glycerol
1 ml of IGEPAL CA-630
0.2 ml of 0.5 M EDTA
0.5 ml of 0.2 M Na3VO4
0.1 ml of 25 mM GTP stock solution (to maintain RAB21 in its activated state)
Stored at -20 °C
Acknowledgments
The RAB21 activity assay was adapted from previously published studies (Franco et al., 2014; Mai et al., 2011; Zhu et al., 2007) and was performed in (Jean et al., 2015). This work was supported by FRSQ, AHA and CRS postdoctoral fellowships to SJ, and NIH RO1 GM078176 and support from the SDCSB NIH P50 GM085764 to AAK.
References
- Franco, I., Gulluni, F., Campa, C. C., Costa, C., Margaria, J. P., Ciraolo, E., Martini, M., Monteyne, D., De Luca, E., Germena, G., Posor, Y., Maffucci, T., Marengo, S., Haucke, V., Falasca, M., Perez-Morga, D., Boletta, A., Merlo, G. R. and Hirsch, E. (2014). PI3K class II alpha controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev Cell 28(6): 647-658.
- Jean, S., Cox, S., Nassari, S. and Kiger, A. A. (2015). Starvation-induced MTMR13 and RAB21 activity regulates VAMP8 to promote autophagosome-lysosome fusion. EMBO Rep 16(3): 297-311.
- Mai, A., Veltel, S., Pellinen, T., Padzik, A., Coffey, E., Marjomaki, V. and Ivaska, J. (2011). Competitive binding of Rab21 and p120RasGAP to integrins regulates receptor traffic and migration. J Cell Biol 194(2): 291-306.
- Zhu, G., Chen, J., Liu, J., Brunzelle, J. S., Huang, B., Wakeham, N., Terzyan, S., Li, X., Rao, Z., Li, G. and Zhang, X. C. (2007). Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5. EMBO J 26(14): 3484-3493.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Jean, S. and Kiger, A. A. (2016). RAB21 Activity Assay Using GST-fused APPL1. Bio-protocol 6(4): e1738. DOI: 10.21769/BioProtoc.1738.
- Jean, S., Cox, S., Nassari, S. and Kiger, A. A. (2015). Starvation-induced MTMR13 and RAB21 activity regulates VAMP8 to promote autophagosome-lysosome fusion. EMBO Rep 16(3): 297-311.
Category
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











