- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of the Volume and Surface Area of Symbiosomes and Vacuoles of Infected Cells in Root Nodules of Medicago truncatula
Published: Vol 5, Iss 23, Dec 5, 2015 DOI: 10.21769/BioProtoc.1665 Views: 11271
Reviewed by: Arsalan DaudiRenate WeizbauerAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2322 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views
Abstract
Legumes are able to form endosymbiotic interactions with nitrogen-fixing rhizobia. Endosymbiosis takes shape in formation of a symbiotic organ, the root nodule. Medicago truncatula (M. truncatula) nodules contain several zones representing subsequent stages of development. The apical part of the nodule consists of the meristem and the infection zone. At this site, bacteria are released into the host cell from infection threads. Upon release, bacteria are surrounded by a host cell-derived membrane to form symbiosomes. After release, rhizobia grow, divide, and gradually colonize the entire host cell of the fixation zone of root nodules. Therefore, mature infected cells contain thousands of symbiosomes, which remain as individual units among other organelles. Visualization of the organization and dynamics of the symbiosomes as well as other organelles in infected cells of nodules is essential to understand mechanisms regulating the development of endosymbiosis between plants and rhizobia. To examine this highly dynamic developmental process, we designed a useful imaging technique that is based on confocal scanning microscopy combined with different fluorescent dyes and GFP-tagged proteins (Gavrin et al., 2014). Here, we describe a protocol for microscopic observation, 3D rendering, and volume/area measurements of symbiosomes and other organelles in infected cells of M. truncatula root nodules. This protocol can be applied for monitoring the development of different host-microbe interactions whether symbiotic or pathogenic.
Keywords: SymbiosisMaterials and Reagents
- Microscope cover glasses
- Microscope slides
- Perlite (Maasmond-Westland, The Netherlands)
- 7 days old seedlings of M. truncatula (seeds can be obtained at The Samuel Roberts Noble Foundation)
- Agrobacterium rhizogenes MSU440 strains (can be obtained at the Laboratory of Molecular Biology, Wageningen University, The Netherlands) containing pUB:GFP-MtVTI11 or pLB:GFP-MtVTI11 binary vector (see Note 1)
- Sinorhizobium meliloti 2011 (see Note 2)
- 0.1 M sodium phosphate (Sigma-Aldrich, catalog numbers: S8282 and S7907 ) buffer (pH 7.2) with 3% sucrose (Sigma-Aldrich, catalog number: S7903). For preparation protocol see Sambrook and Russell (2001)
Note: Sodium phosphate monobasic (Sigma-Aldrich, catalog number: S8282) and Sodium phosphate dibasic (Sigma-Aldrich, catalog number: S7907) - 10 μg/ml Propidium iodide (store at 4 °C) (Sigma-Aldrich, catalog number: P4170 )
- 1% paraformaldehyde (Electron Microscopy Sciences, catalog number: 15700 )
- 0.75% glutaraldehyde (Electron Microscopy Sciences, catalog number: 16000 )
- 50 mM phosphate buffer (Sambrook and Russell, 2001)
- Fixative (see Recipes)
Equipment
- Fluorescence stereomicroscope (Leica Microsystems, model: MZFLIII )
- Confocal laser scanning microscope with digital camera (ZEISS, model: Axiovert 100M )
- Razor blades
- Fine tweezers (Structure Probe, Dumont, model: 0T05B-XD )
Software
- IMARIS software (Bitplane) with modules for data management, visualization, 3D rendering and analysis
- Zeiss LSM Image Browser
Procedure
- Generation of transgenic nodules
- Generation of pUB:GFP-MtSYP22 or pLB:GFP-MtSYP22 expressing roots by Agrobacterium rhizogenes1-mediated transformation of 7 days old seedlings of M. truncatula. For transformation protocol see Limpens et al. (2004). Plants were grown for 14 days after transformation.
- Inoculation of composite M. truncatula plants with pUB:GFP-MtSYP22 or pLB:GFP-MtSYP22 transgenic roots with Sinorhizobium meliloti (S. meliloti) 2011 (Sambrook and Russell, 2001). Composite Medicago plants with transgenic roots are transferred to perlite saturated with Färhaeus medium without Ca(NO3)2 (Limpens et al., 2004). After 2 days, plants are inoculated with 2 ml of the S. meliloti 2011 culture (OD600 nm of 0.1) per plant and grown for 2 weeks (21 °C; 16/8 h light/darkness).
- Generation of pUB:GFP-MtSYP22 or pLB:GFP-MtSYP22 expressing roots by Agrobacterium rhizogenes1-mediated transformation of 7 days old seedlings of M. truncatula. For transformation protocol see Limpens et al. (2004). Plants were grown for 14 days after transformation.
- Sample preparation for image acquisition
- Transgenic nodules (14 days after inoculation) are selected using a fluorescent stereomicroscope with DsRED1 filter settings (Figure 1A-B).
- Harvested nodules are collected with tweezers in an Eppendorf tube with 500 µl of fixative for 1 h fixation at 4 °C.
- Fixed nodules are hand-sectioned by razor blades in phosphate buffer (Figure 1C). Try to do sections as thin as possible.
- Nodule sections are collected by a tweezer in an Eppendorf tube with propidium iodide solution for 10-20 min staining at room temperature.
- Place nodule sections in a drop of phosphate buffer on a microscopic glass slide. Then place an edge of the cover glass over the sample and carefully lower the cover; make sure there is no air between tissue and glass.
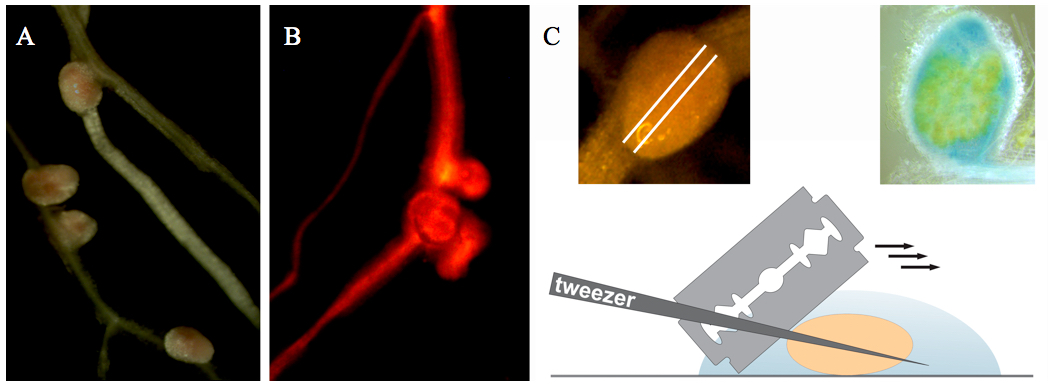
Figure 1. Collection and sectioning of Medicago root nodules. A. Medicago root nodules. B. Medicago root nodules on a transgenic root (selected on the basis of red fluorescence resulting from DsRED1 expression). C. Nodule sectioning.
- Transgenic nodules (14 days after inoculation) are selected using a fluorescent stereomicroscope with DsRED1 filter settings (Figure 1A-B).
- Image acquisition
- Observe the nodule section under the confocal microscope using an oil immersion objective lens (x63 or higher) with 488 nm excitation and 505-530 nm emission wavelengths for GFP and with 543 nm excitation and 636 nm emission wavelengths for propidium iodide. Filters: Ch1: LP560, Ch2: BP505-530. Pinhole: 106 μm.
- Find a group of infected cells at the developmental stage of interest and focus on it.
- Obtain Z-stack serial images of region of interest. Z-stack size: 512 x 512 x 20-73 pixel. Number of single images in Z-stacks 20-67 depending on the position and size of scanned cells. Using Projection tool you can create a 3D projection image or a rotation in 3D, which can be animated and exported as an AVI movie (Figure 2, Video 1).
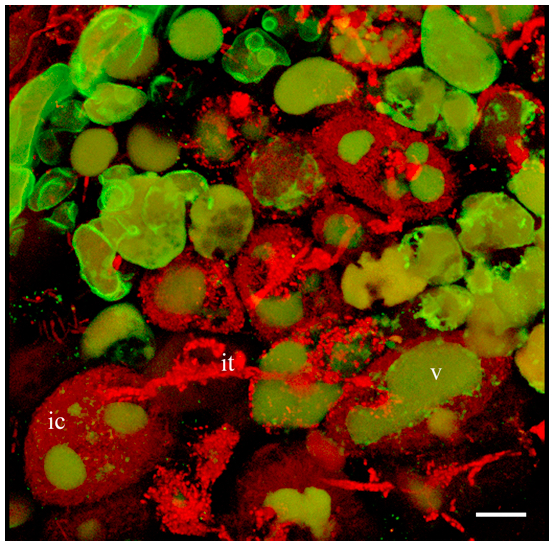
Figure 2. 3D projection of Z-stack serial images of nodule cells expressing GFP-MtSYP22 (green). Bacteria and bacteroids are contrasted by propidium iodide (red). ic, infected cell; it, infection thread; v, vacuole. Scale bar 40 µm.Video 1. 3D projection of nodule cell
- Observe the nodule section under the confocal microscope using an oil immersion objective lens (x63 or higher) with 488 nm excitation and 505-530 nm emission wavelengths for GFP and with 543 nm excitation and 636 nm emission wavelengths for propidium iodide. Filters: Ch1: LP560, Ch2: BP505-530. Pinhole: 106 μm.
- Image processing: 3D rendering and volume/area measurements of vacuoles and symbiosomes in infected cells
- Open Z-stack serial images of the nodule using IMARIS software.
- Select a region of interest by 3D Crop operation for further manipulations (Figure 3).
- If an image is too dim to view, adjust brightness of channels individually in the Display Adjustment window.
- To get rid of out of focus light or other artefacts before measurements, select Smoothing in the Image Processing menu. Select the channels you want to have filtered and filter out noise signal from the 3D image.
- Generate surfaces of vacuoles and symbiosomes of infected cells in the Surface generation wizard (select the Source Channel for segmentation: for vacuoles pick GFP channel and for symbiosomes RFP channel). To generate the surface of the whole infected cell, use manual 3D construction mode.
- Adjust the threshold (the thresholded regions are shown in grey during adjustment).
- Generated and complete surfaces will be present in the image with changed colours (Figure 3B). You can take measurements from your surfaces by pressing the measure icon (Imaris Measurement). If necessary, adjust the measurement parameters and use filters. Using Movie Maker you can create animations with generated surfaces (Video 2).
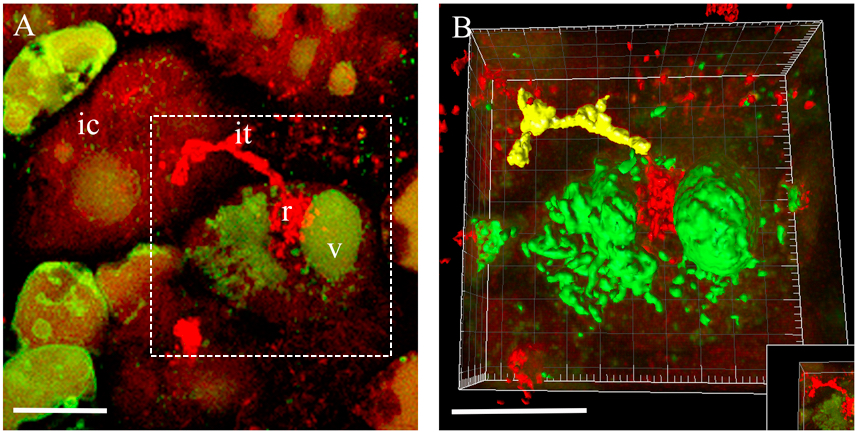
Figure 3. 3D rendering of the infected cell at the moment of bacteria release in Medicago root nodules. A. 3D projection of Z-stack serial images of nodule cells expressing at the moment of bacteria release. B. An example of 3D rendering of the infected cell outlined in (A). ic, infected cell; it, infection thread; r, release of bacteria; v, vacuole. Scale bar 20 µm.Video 2. Fragmented vacuole of an infected cell
- Open Z-stack serial images of the nodule using IMARIS software.
Notes
- MtSYP22 is a vacuole SNARE protein, controlling pre-vacuolar compartment-to-vacuole trafficking. GFP-MtSYP22 fusion protein allows visualizing vacuoles in nodule cells. pUB:GFP-MtSYP22 fusion is controlled by a ubiquitin promoter expressed in the apical part of root nodules (meristem and the infection zone). pLB:GFP-MtSYP22 fusion is driven by a leghemoglobin promoter, which is active in mature infected cells of the fixation zone of root nodules. These binary vectors enable the selection of transgenic roots and nodules based on additional DsRED1 (Discosoma sp. red fluorescent protein) expression.
- Sinorhizobium meliloti 2011 is a symbiont of M. truncatula (can be obtained at Laboratory of Molecular Biology, Wageningen University).
Recipes
- Fixative
1% paraformaldehyde
0.75% glutaraldehyde
50 mM phosphate buffer (Sambrook and Russell, 2001)
Stored at -20 °C
Acknowledgments
We thank our colleagues Norbert de Ruijter for assistance with confocal imaging and T. W. J. Gadella (University of Amsterdam) for help with the analysis of Imaris 3D reconstructed images. A. G. received a Ph.D. fellowship from the EPS School of Biological Sciences (Wageningen University).
References
- Gavrin, A., Kaiser, B. N., Geiger, D., Tyerman, S. D., Wen, Z., Bisseling, T. and Fedorova, E. E. (2014). Adjustment of host cells for accommodation of symbiotic bacteria: Vacuole defunctionalization, HOPS suppression, and TIP1g retargeting in Medicago. Plant Cell 26(9): 3809-3822.
- Limpens, E., Ramos, J., Franken, C., Raz, V., Compaan, B., Franssen, H., Bisseling, T. and Geurts, R. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55(399): 983-992.
- Sambrook, J. and Russell, D. W. (2001). Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, third edition.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gavrin, A. and Fedorova, E. E. (2015). Quantification of the Volume and Surface Area of Symbiosomes and Vacuoles of Infected Cells in Root Nodules of Medicago truncatula. Bio-protocol 5(23): e1665. DOI: 10.21769/BioProtoc.1665.
Category
Plant Science > Plant physiology > Endosymbiosis
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










