- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Secretion Assay in Shigella flexneri
Published: Vol 4, Iss 22, Nov 20, 2014 DOI: 10.21769/BioProtoc.1302 Views: 11612
Reviewed by: Fanglian HeKanika Gera

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

From Llama to Nanobody: A Streamlined Workflow for the Generation of Functionalised VHHs
Lauren E.-A. Eyssen [...] Raymond J. Owens
Mar 20, 2024 6244 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2095 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2171 Views
Abstract
Shigella flexneri (S. flexneri) is a Gram-negative bacterium that causes gastroenteritis and shigellosis in humans. In order to establish and maintain an infection, S. flexneri utilises a type three secretion system (T3SS) to deliver virulence factors called effector proteins into the cytoplasm of host cells, facilitating e.g. uptake into the host cell and escape from the endosome. Secretion through the T3SS is tightly regulated and is usually triggered by host-cell contact, but can also be artificially stimulated in vitro. In this assay, the dye Congo red is used to induce T3SS-dependent secretion of S. flexneri (Parsot et al., 1995) and secreted proteins are concentrated from the culture supernatant by precipitation with trichloroacetic acid. The assay presented here can easily be adapted to the secretion analysis of other bacteria utilising a T3SS, such as Salmonella typhimurium, which constitutively secrete when grown at 37 °C (Collazo et al., 1995; Pegues et al., 1995), or pathogenic species of Yersinia, where secretion can be induced by calcium deprivation (Heesemann et al., 1986; Forsberg et al., 1987).
Keywords: Shigella flexneriMaterials and Reagents
- Tryptone soy agar (TSA) (Carl Roth, catalog number: CP70.1 )
- Shigella flexneri (strain M90T) on a Congo red tryptone soy agar plate
- 99% Trichloroacetic acid (TCA) (Carl Roth, catalog number: 8789 )
- Acetone (Merck KGaA, catalog number: 1000142511 )
- TRIZMA base (Sigma-Aldrich, catalog number: T1503 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771 )
- Bromophenol blue (Sigma-Aldrich, catalog number: B5525 )
- Glycerol (Sigma-Aldrich, catalog number: G7757 )
- 2-Mercaptoethanol (Sigma-Aldrich, catalog number: M3148 )
- LB medium (Carl Roth, catalog number: X968.2 ) (see Recipes)
- Tryptone soy agar plates (see Recipes)
- Sample buffer (see Recipes)
- Congo red stock solution (Sigma-Aldrich, catalog number: C6767 ) (see Recipes)
Note: Used as 5 mg/ml stock solution.
Equipment
- 5 ml syringes (Henke-Sass, Wolf, catalog number: 5050.000V0 )
- 0.2 µm syringe filters (GE Healthcare, catalog number: 10462200 )
- Culture tubes
- 50 ml Erlenmeyer flasks
- 1.5 ml microtubes
- Microfuge
- 37 °C shaking incubator
- Laminar flow hood
- Spectrophotometer
Procedure
- Prepare a pre-culture by inoculating 2 ml of LB medium with a single colony of S. flexneri from a Congo red tryptone soy agar plate and grow overnight (~16 h) at 37 °C with agitation (180 rpm).
Note: Ensure that the colonies used are secretion-competent, i.e. forming red colonies on the plate due to Congo red absorption (Payne and Finkelstein, 1977).
- Subculture by diluting 100 µl of the pre-culture in 10 ml of fresh LB medium (i.e. 1:100 dilution).
- Grow to OD600 (absorbance at 600 nm) of 0.3-0.4 at 37 °C with agitation.
Note: In case genes from expression plasmids are to be induced, this should be done at OD600 of ~0.1.
- Add Congo red to a final concentration of 200 µg/ml (i.e. 400 µl of 5 mg/ml stock solution to 10 ml of culture).
- Grow for 2-3 h at 37 °C with agitation – the final OD600 of the culture should be between 2 and 3.
- Pellet cells by centrifugation for 10 min at 10,000 x g and 4 °C.
Note: For the following TCA precipitation a supernatant volume corresponding to 1 ml culture of OD600=2 is used, meaning that centrifugation of 1.4 ml of culture in 1.5 ml tubes should be sufficient (considering the dead volume of the filter units in step 7).
- Transfer the supernatant to the 5 ml syringes and filter through 0.2 µm filters.
Note: For maximum yield remove the piston from the syringe and add the filter. Transfer the supernatant into the syringe and replace the piston- the additional air will minimize the volume lost in the filter.
- Take a volume of filtered supernatant corresponding to 1 ml culture of OD600=2 and add ice-cold 99% trichloroacetic acid to a final concentration of 10% (v/v).
Note: Congo red will turn blue at this point and precipitate as well.
- Transfer the tubes to -20 °C for 20 min.
- Centrifuge at top speed (12,000 x g to 16,000 x g) in a microfuge for 30 min at 4 °C.
- Carefully discard the supernatant and rinse the pellet with 1 ml ice-cold acetone.
- Centrifuge again (12,000 x g to 16,000 x g, 30 min, 4 °C).
- Carefully remove the supernatant and let the pellet air dry for 15 min.
- Resuspend the pellet in sample buffer.
Note: The volume required may vary with the method of detection after electrophoresis: for analysis by Coomassie staining, resuspension in 25 µl to 100 µl will produce sufficiently strong bands.
- Analyse by SDS-PAGE through 12% acrylamide gels.
Representative data
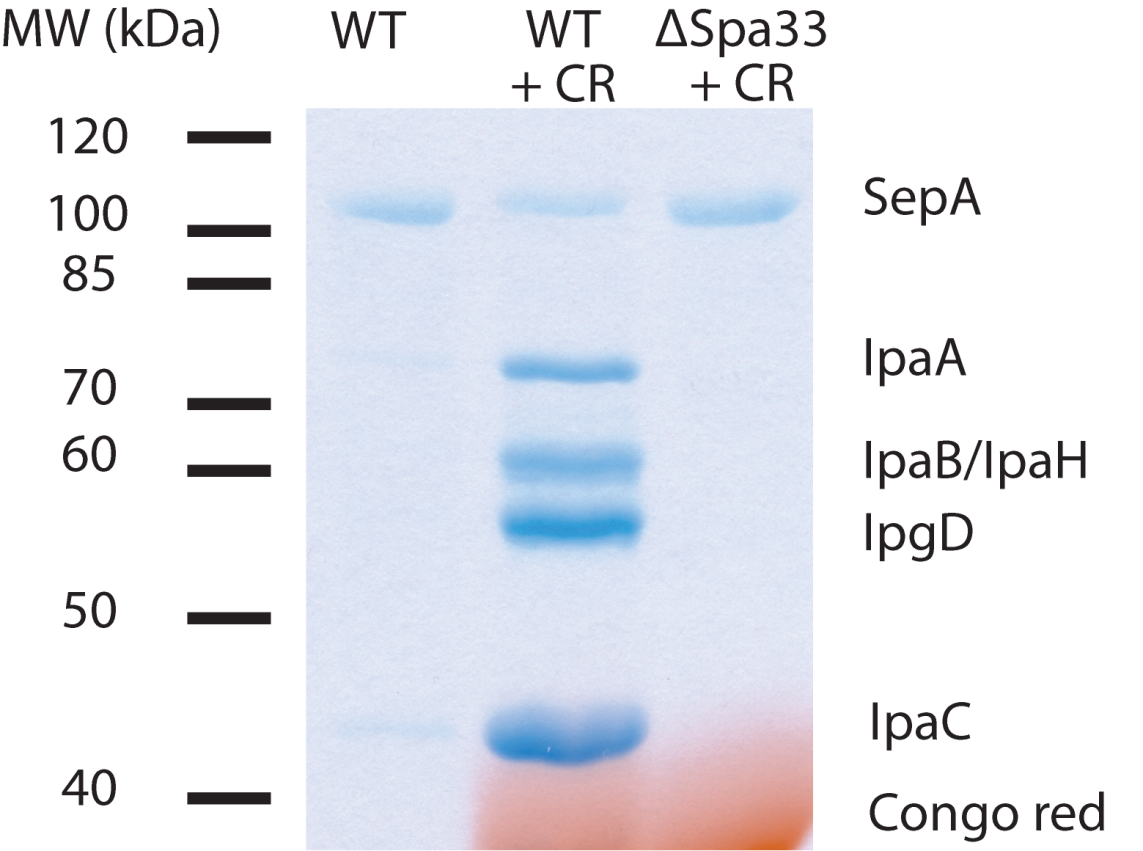
Figure 1. Example of secretion from S. flexneri. TCA-precipitated pellets were resuspended in 25 µl of sample buffer and 20 µl loaded on a 12% polyacrylamide gel. CR = Congo red. ΔSpa33 is a T3SS-deficient mutant. IpaA, IpaB, IpaH, IpgD and IpaC are effector proteins. SepA is a T3SS-independent secreted protein and serves as an intrinsic loading control.
Note: IpaD may be discernible at 37 kDa, but is often obscured by Congo red. Congo red will return to its original color by soaking the gel in 100 mM Tris (pH 9.0) after Coomassie staining.
Recipes
- LB medium
25 g LB medium powder
Water to 1 L
Note: It may be necessary to adjust the pH to 7.0-7.5 with a few drops of 1 M NaOH solution. This is usually not required, since LB medium is weakly buffered in this pH range.
Autoclave the solution
- Tryptone soy agar plates
20 g Tryptone soy agar powder
0.05 g Congo red powder
Water to 500 ml
Note: It may be necessary to adjust the pH to 7.0-7.5 with a few drops of 1 M NaOH solution. This is usually not required, since TSA is weakly buffered in this pH range.
Autoclave the solution
Cool to ~50 °C and pour plates
- Sample buffer
SDS-loading buffer with increased buffer capacity
10% glycerol (v/v)
360 mM Tris-HCl (pH 6.8)
2% SDS (w/v)
1.25% 2-mercaptoethanol (v/v)
0.01% bromophenol blue (w/v)
- Congo red stock solution
5 mg of Congo red powder per ml water
Note that the dye content in the Congo red powder used here is ~40% and may differ between manufacturers. The stock solution is stable at room temperature for several months. When kept in the fridge or at -20 °C, Congo red may precipitate. Warm the solution to 37 °C to re-dissolve.
Acknowledgments
This work has been financially supported by the European Research Council under the European Community's Seventh Framework Programme (FP7/2007-2013) and a Ph.D. Scholarship from the International Max Planck Research School for Infectious Diseases and Immunology.
The protocol presented here has been adapted from Dohlich et al. (2014).
References
- Collazo, C. M., Zierler, M. K. and Galan, J. E. (1995). Functional analysis of the Salmonella typhimurium invasion genes invl and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol Microbiol 15(1): 25-38.
- Dohlich, K., Zumsteg, A. B., Goosmann, C. and Kolbe, M. (2014). A substrate-fusion protein is trapped inside the Type III Secretion System channel in Shigella flexneri. PLoS Pathog 10(1): e1003881.
- Forsberg, A., Bolin, I., Norlander, L. and Wolf-Watz, H. (1987). Molecular cloning and expression of calcium-regulated, plasmid-coded proteins of Y. pseudotuberculosis. Microb Pathog 2(2): 123-137.
- Heesemann, J., Gross, U., Schmidt, N. and Laufs, R. (1986). Immunochemical analysis of plasmid-encoded proteins released by enteropathogenic Yersinia sp. grown in calcium-deficient media. Infect Immun 54(2): 561-567.
- Parsot, C., Menard, R., Gounon, P. and Sansonetti, P. J. (1995). Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol Microbiol 16(2): 291-300.
- Payne, S. M. and Finkelstein, R. A. (1977). Detection and differentiation of iron-responsive avirulent mutants on Congo red agar. Infect Immun 18(1): 94-98.
- Pegues, D. A., Hantman, M. J., Behlau, I. and Miller, S. I. (1995). PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol 17(1): 169-181.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Reinhardt, J. and Kolbe, M. (2014). Secretion Assay in Shigella flexneri. Bio-protocol 4(22): e1302. DOI: 10.21769/BioProtoc.1302.
- Dohlich, K., Zumsteg, A. B., Goosmann, C. and Kolbe, M. (2014). A substrate-fusion protein is trapped inside the Type III Secretion System channel in Shigella flexneri. PLoS Pathog 10(1): e1003881.
Category
Microbiology > Microbe-host interactions > Bacterium
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link