- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2025
Volume: 15, Issue: 20
Cancer Biology
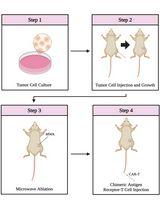
Combining Microwave Ablation With CAR-T-Cell Therapy in Tumor-Bearing Mouse Models
Cell Biology
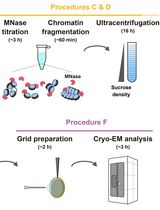
Preparation of Chromatin Fragments From Human Cells for Cryo-EM Analysis
Studying Cargo Transport Using RudLOV
Generation of Insulin-Producing Alpha TC1-6 Cells Using EpiCRISPR System for Targeted DNA Methylation
Immunology
High-Dimensional Phospho-CyTOF Characterization of T-Cell Activation Responses in Whole Blood
Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Molecular Biology
SunTag-Based Single-Molecule Translation Imaging in Caenorhabditis elegans
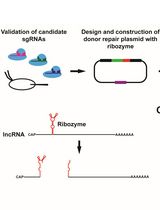
Ribozyme-Mediated Knockdown of lncRNA Gene Expression in Drosophila
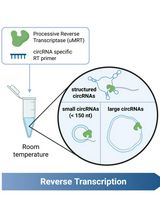
Efficient circRNA Detection Using the Processive Reverse Transcriptase uMRT
Plant Science
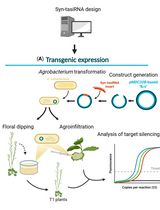
Effective Gene Silencing in Plants by Synthetic Trans-Acting siRNAs Derived From Minimal Precursors
Direct Plant Regeneration From Immature Male Inflorescence of Banana (Musa spp.)
Stem Cell
Rapid and Simplified Induction of Spinal Motor Neurons From Human Induced Pluripotent Stem Cells