- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2025
Volume: 15, Issue: 17
Biochemistry
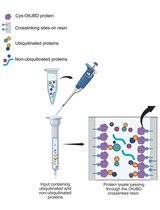
Use of a High-Affinity Ubiquitin-Binding Domain to Detect and Purify Ubiquitinated Substrates and Their Interacting Proteins
Bioinformatics and Computational Biology
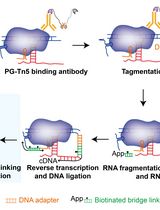
Simultaneous Capture of Chromatin-Associated RNA and Global RNA–RNA Interactions With Reduced Input Requirements
Biophysics
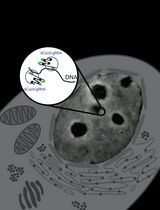
Real-Time Imaging of Specific Genomic Loci With CRISPR/dCas9 in Human Cells Using CRISPRainbow
Cancer Biology
NanoPDLIM2-Based Combination Therapy for Lung Cancer Treatment in Mouse Preclinical Studies
Immunology
Quantitative Microscopy for Cell–Surface and Cell–Cell Interactions in Immunology
Novel Experimental Approach to Investigate Immune Control of Vascular Function: Co-culture of Murine Aortas With T Lymphocytes or Macrophages
Neuroscience
Ultrafast Isolation of Synaptic Terminals From Rat Brain for Cryo-Electron Tomography Analysis
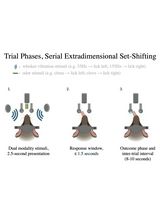
Training Mice to Perform Attentional Set-Shifting Under Head Restraint
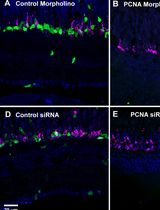
Efficient Gene Knockdown in Adult Zebrafish Retina by Intravitreal Injection
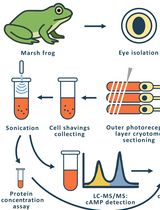
Time-Resolved cAMP Level Determination in Frog Retina Samples Using LC–MS/MS
Constructing and Implementing a Low-Cost On-Demand Morris Water Maze Platform
Plant Science
PhosphoLIMBO: An Easy and Efficient Protocol to Separate and Analyze Phospholipids by HPTLC From Plant Material
New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage
Stem Cell
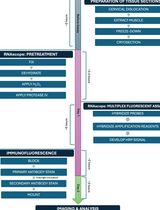
Simultaneous RNA Fluorescent In Situ Hybridization and Immunofluorescent Staining of Mouse Muscle Stem Cells on Fresh Frozen Skeletal Muscle Sections